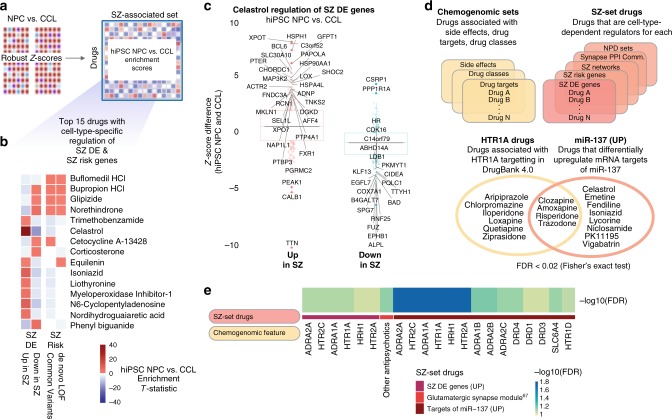
Fig. 4.
Differences in drug-induced responses between hiPSC NPCs and CCLs. a A gene-set enrichment approach was applied to identify drugs that regulated expression of SZ-set genes differently in profiles generated from hiPSC NPCs and CCLs. b Top 15 drugs with highest collective cell-type-specific perturbation of genes differentially expressed in prefrontal cortex of individuals with SZ (SZ DE), and/or genes harboring common and de novo SZ-risk-associated variants. Squares with enrichment FDR > 0.1 are shown as white. c Celastrol showed strong hiPSC NPC/CCL differential regulation of SZ DE genes (absolute Z-score difference > 3 labeled as solid filled circles with dark red or blue color). d To identify pharmacological features associated with drugs that differentially perturbed SZ sets in hiPSC NPCs compared with CCLs, we performed a chemogenomic enrichment analysis. SZ sets that were differentially perturbed by at least three drugs in the study (SZ-set drugs) were identified and enrichment testing was performed to identify over-represented drug targets, drug classes, and side effects. Drugs that differentially perturbed SZ sets between hiPSC NPCs and CCLs shared two pharmacological features: targeting of the serotonin receptor HTR1A and upregulation of the SZ-associated microRNA miR-137. e Chemogenomic features that were over-represented among drugs that differentially regulated various SZ sets depending on cell type (hiPSC NPC vs. CCL) context. Strong enrichment observed for drugs that differentially regulated SZ DE genes and targets of miR-137, particularly by antipsychotic drugs that targeted adrenergic, serotonergic, and dopaminergic neurotransmitter receptors (chemogenomic features). Abbreviations: SZ DE, genes differentially expressed in SZ; SZ Risk, genes harboring SZ-risk-associated loci; SZ Networks, SZ-associated coexpression networks; Synapse PPI Communities, NPD enriched synaptic protein seeded clique communities; and NPD Sets, diverse NPD-associated sets, e.g., miR-137 targets