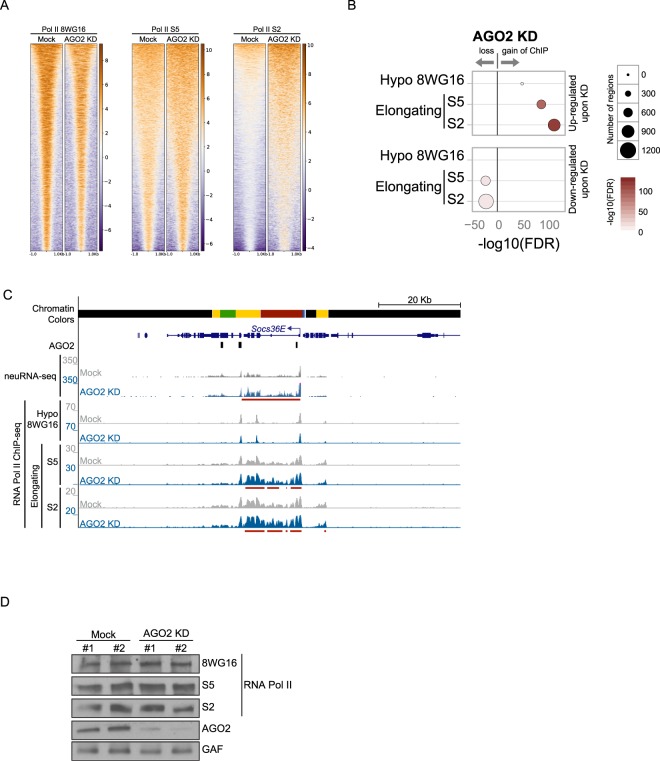
Figure 3.
AGO2 reduces Pol II elongation. (A) Heatmaps of Pol II 8WG16, Pol II S5 and Pol II S2 peaks in mock-treated cells or upon AGO2 depletion sorted by decreasing average ChIP-seq signal in respective mock-treated cells. The horizontal axis corresponds to distance from peak center for each analyzed factor. (B) Differential ChIP-seq analysis for hypophosphorylated (8WG16) and elongating (S5 and S2) Pol II forms on neuRNA-seq affected genes upon depletion of AGO2. Size of circles indicates the number of total genes across the genome that display differential binding (see text for intersection values), and color indicates −log10(FDR) where FDR is the p-value from FETs adjusted for multiple comparisons. Only statistically significant results are shown for clarity. (C) Screenshot showing the example gene Socs36E, which is up-regulated in AGO2 knockdown. AGO2 ChIP peaks (black bars) and ChIP-seq signal of hypophosphorylated, S5, and S2 forms of Pol II are shown. Red bars below signal tracks correspond to statistically significant increases in neuRNA-seq or ChIP-seq signal relative to mock sample. (D) Western blot analysis showing no changes in Pol II levels in AGO2 knockdown. AGO2 is efficiently knocked down, and GAF is included as loading control. The number above each lane indicates biological replicate. Representative western blots (cropped) are shown.