Abstract
In shrimp aquaculture, farming systems are carefully managed to avoid rearing failure due to stress, disease, or mass mortality, and to achieve optimum shrimp production. However, little is known about how shrimp farming systems affect biogeochemical parameters and bacterial communities in rearing water, whether high stocking densities (intensive system) will increase the abundance of pathogenic bacteria. In this study, we characterized bacterial communities in shrimp ponds with different population densities. Water quality, such as physical parameters, inorganic nutrient concentrations, and cultivable heterotrophic bacterial abundances, including potential pathogenic Vibrio, were determined in moderate density/semi-intensive (40 post-larvae m-3) and high density/intensive shrimp ponds (90 post-larvae m-3), over the shrimp cultivation time. Free-living and particle-attached bacterial communities were characterized by amplicon sequencing of the 16S rRNA gene. Suspended particulate matter (SPM), salinity, chlorophyll a, pH, and dissolved oxygen differed significantly between semi-intensive and intensive systems. These variations contrasted with the equal abundance of cultivable heterotrophic bacteria and inorganic nutrient concentrations. Bacterial communities were dominated by Gammaproteobacteria, Alphaproteobacteria, Flavobacteriia, Bacilli, and Actinobacteria. Halomonas and Psychrobacter were the most dominant genera in the particle-attached fractions, while Salegentibacter, Sulfitobacter, and Halomonas were found in the free-living fractions of both systems. Redundancy analysis indicated that among the observed environmental parameters, salinity was best suited to explain patterns in the composition of both free-living and particle-attached bacterial communities (R2: 15.32 and 12.81%, respectively), although a large fraction remained unexplained. Based on 16S rRNA gene sequences, aggregated particles from intensive ponds loaded a higher proportion of Vibrio than particles from semi-intensive ponds. In individual ponds, sequence proportions of Vibrio and Halomonas displayed an inverse relationship that coincided with changes in pH. Our observations suggest that high pH-values may suppress Vibrio populations and eventually pathogenic Vibrio. Our study showed that high-density shrimp ponds had a higher prevalence of Vibrio, increased amounts of SPM, and higher phytoplankton abundances. To avoid rearing failure, these parameters have to be managed carefully, for example by providing adequate feed, maintaining pH level, and removing organic matter deposits regularly.
Keywords: Litopenaeus vannamei, Illumina sequencing, pathogenic bacteria, aggregates, salinity, Indonesia
Introduction
Litopenaeus vannamei (Boone, 1931), white-leg shrimp, is an important species in aquaculture industry, which is widely reared in subtropical to tropical regions. They grow rapidly, have high survival rates even at high densities, possess a wide tolerance range of salinity and temperature, and can be cultivated in indoor (tanks or recirculating aquaculture systems) or outdoor facilities (ponds; Cuzon et al., 2004). Anderson et al. (2016) reported that world shrimp production in 2015 was 4.2 million tons of which L. vannamei contributed 75%. However, severe economic loss due to massive shrimp mortality has been occurring since 1987. For the period 1987–1994, economic loss reached USD 3 billion, representing 40% of the total production capacity of the industry (Brun et al., 2009; Walker and Winton, 2010). Moreover, a recent emerging bacterial disease, known as acute hepatopancreatic necrosis disease (AHPND), has occurred in Asian and Mexican shrimp aquaculture, causing an annual loss amounting to USD 1 billion with shrimp mortality exceeding 70% (Global Aquaculture Alliance [GAA], 2013, De Schryver et al., 2014; Soto-Rodriguez et al., 2015).
The intensification of the shrimp industries resulted in changes of farming systems and sustainability. Until the year 2000, 70% of shrimp farming in Indonesia was conducted in extensive pond systems (Kautsky et al., 2000), but recently changed to semi-intensive or intensive systems (Kementerian Kelautan dan Perikanan [KKP], 2015). Different shrimp stocking densities affect rearing processes, including a defined nutritional input and shrimp production. Robertson et al. (1993) reported that the shrimp growth rate increased progressively as feeding frequency increased from 1 to 4 times per day. However, only a portion of the nutrients in the feed is consumed, assimilated, and retained as shrimp biomass. Shrimps only incorporate 24 to 37% of nitrogen and 11 to 20% of phosphorus from the feed into their bodies (Funge-Smith and Briggs, 1998; Sahu et al., 2013). In addition, 15% of nitrogen losses occurs during the first 2 h of immersion of feed pellets into the pond water (Smith et al., 2002). These unused nutrients will lead to a change of pH and dissolved oxygen (DO) in the water column and pond sediment, eutrophication, proliferation of bacteria and plankton, and an increase of particulate organic matter (Avnimelech et al., 1994; Martin et al., 1998). Furthermore, Kautsky et al. (2000) reported that the risk of shrimp diseases often increased with culture intensity and high stocking densities. Thus, shrimp diseases, along with bad water quality management, might threaten shrimp farming sustainability.
Free-living (FL) and particle-associated (PA) bacterial communities from the same water sample can be distinct (Bidle and Fletcher, 1995; Riemann and Winding, 2001; Jain and Krishnan, 2017; Yang et al., 2017). As organic-rich particles, aggregates provide a suitable habitat for microorganisms to take up nutrients, and shelter from predators, as well as from destructive physical factors (Lyons et al., 2010; Kramer et al., 2013). In addition, aggregates can accommodate higher bacterial abundance and diversity than the adjacent water column (Kiørboe et al., 2003). Due to water movement in ponds, organic matter agglomerates and forms large flocs or aggregates (Hargreaves, 2013; Avnimelech, 2014), which might facilitate bacterial settlement and proliferation. Previous studies about bacterial community compositions (BCCs) in shrimp pond waters have yet to analyze FL and PA fractions separately (Sombatjinda et al., 2011; Zhang D. et al., 2014; Xiong et al., 2016; Hou et al., 2017), resulting in a paucity of information on BCC in both fractions.
Although the causative agents for bacterial diseases in shrimps have been identified (Lavilla-Pitogo et al., 1998; Kharisma and Manan, 2012; Soto-Rodriguez et al., 2015; Xiao et al., 2017), preventive efforts to minimize disease outbreaks seem to still be ineffective. Studies in temperate ecosystems showed that pathogenic bacteria have been found in aggregates (Lyons et al., 2005; Froelich et al., 2013). Based on that evidence, we propose that particle abundance can be used to estimate the potential proliferation of pathogenic bacteria in shrimp farming, and that controlling aggregates may become an effective tool to manage the spread and survival of pathogens. Therefore, it is necessary to investigate water quality parameters, bacterial abundance, and BCC from different shrimp farming systems for both FL and PA fractions, over shrimp cultivation time.
The current study aims to comprehend the effect of shrimp farming systems of different intensity on water quality parameters and BCC, to elucidate factors affecting bacterial communities, and to evaluate the presence of pathogenic bacteria, including pathogenic V. parahaemolyticus in the FL and the PA/aggregates fraction. We hypothesize that (i) different shrimp farming systems affect pond water quality, including suspended particulate matter (SPM) loading, bacterial abundance and community composition, and (ii) SPM loads more bacterial cells, including pathogenic bacteria.
Materials and Methods
Sample Collection and Sampling Sites
Water samples were collected between 9–10 a.m. from no water-exchange and plastic lining ponds (square shape in size 2700–3000 m2, water depth 1.3–1.5 m) of semi-intensive (40 post-larvae m-3, three ponds) and intensive (90 post-larvae m-3, three ponds) systems, during a cycle of shrimp rearing at day 10, 20, 30, 40, 50, 60, and 70 (September to November 2016). Shrimp ponds were located in Rembang Regency, Central Java, Indonesia (-6°37′41.13″ S 111°30′1″ E and -6°42′11.66″ S 111°21′54″ E, for semi-intensive and intensive system, respectively). Two liters of water were taken from 5 points of each pond at 1 m depth, and then mixed. Two liters of the mixed water were prepared for bacterial abundance and community analysis; 3 liters for SPM and inorganic nutrient analysis. Samples were stored in a cold and dark container, and transported to the laboratory at Diponegoro University, Semarang, Indonesia, for immediate analysis. Remaining water was used for physical parameter measurement. Total harvest of each pond was recorded at the end of shrimp rearing.
Environmental Parameters
Salinity, temperature, pH, chlorophyll a, DO, and turbidity were measured ex situ, using calibrated Manta Eureka 2 multi-probes (Eureka Environmental Engineering, TX, United States).
Of each water sample, 0.3 L were filtered through a 0.45 μm syringe filter and the filtrate was poisoned with 1.2 mL of a 0.35 g L-1 HgCl2 solution for inorganic nutrient analysis (ammonium: NH4+, nitrate: NO3-, nitrite: NO2, phosphate: PO43- and silicate: SiO44-). The samples were stored at -20°C until analysis. Inorganic nutrient measurements were done in triplicates at the Laboratory of Chemistry, Research Center for Oceanography (LIPI), Jakarta, Indonesia, according to the colorimetry method by Strickland and Parsons (1972), with a Shimadzu UV-1800 spectrophotometer.
Suspended particulate matter was measured as dry mass on pre-combusted GF/F filters (porosity 0.7 μm, ø 47 mm, VWR, France) in triplicates after filtration of a known volume of water sample (0.1–0.5 L). Weight of the filters was determined using a precision balance (ME 36S, Sartorius, Göttingen, Germany) after drying the filters for 24 h at 40°C.
Bacterial Abundance
To obtain cultivable heterotrophic bacterial number, 100 μL of water samples (dilution factor 10-2–10-5) were plated onto Marine Agar 2216 (Difco, United States) and incubated at 28°C (room temperature) for 48 h. While potentially pathogenic Vibrio were isolated by inoculating 100 μL of undiluted to 10-4 diluted water sample onto selective Thiosulfate Citrate Bile Salts Sucrose(TCBS) medium (Roth, Karlsruhe, Germany), followed by incubation at 35°C for 24 h.
Samples for total bacterial cell counts were prepared by fixing 50 mL water with 4% v/v paraformaldehyde and stored at 4°C for 24 h. The dilution factors for the samples were as follows: undiluted for day 10 samples, 5 × 10-1 for day 20 and 30 samples, 10-1 for day 40, 50, and 60 samples, and 5 × 10-2 for day 70 samples. Ten milliliters of diluted fixed samples were subsequently filtered through 3.0 and 0.2 μm polycarbonate filters (ø 47 mm and 25 mm, respectively, Whatman, Dassel, Germany) to determine bacterial cells in the particle-attached (PA) and the free-living (FL) fractions, respectively. For the FL fractions at day 30, 40, and 70, only 5 ml filtrates were filtered. Filters were air dried and stored at -20°C for further staining. A 4′,6-diamidino-2-phenylindole (DAPI) staining was performed according to Kepner and Pratt (1994) for selected samples (10, 40, 50, 60, and 70 days samples). Filters were stained with 1 μg mL-1 of DAPI solution for 5 min, then washed in 80% ethanol and rinsed with sterile distillated water. Stained filters were air dried in the dark for 30 min, and then mounted with 10 μL of mounting solution consisting of 3:1 Citifluor AF mounting medium (Citifluor Ltd., London, United Kingdom) and Vectashield (Vector Laboratories Inc., Burlingame, United States). Bacterial cells, as well as size and number of aggregates were observed under a fluorescence microscope Axio Imager.D2 (Zeiss, Jena, Germany) at 1000× magnification. Bacterial cell abundance in FL filters was calculated from 30 photos per filter, using the free software ImageJ. Bacterial cells in PA filters were manually counted from 10 aggregates per filter of similar size. Sizes of aggregates were determined using a net micrometer grid (12.5 mm × 12.5 mm, divided into 10 × 10 fields, which is equal to 15,625 μm2 at 1000× magnification). The cell average per filter was then divided by volume of filtered samples multiplied by dilution and factor of effective filter area (which was 31888 at magnification 1000×, for a filtration funnel with a diameter 25 mm, Millipore, Darmstadt, Germany) and number of aggregates (4 aggregate sizes per filter, Supplementary Table 1), for the FL and the PA fraction, respectively.
Molecular Analysis of Bacterial Communities
Five hundred milliliters of water samples were filtered subsequently through 3.0 and 0.2 μm polycarbonate filters (ø 47 mm, Whatman, Dassel, Germany) for PA and FL bacterial fractions, respectively. Genomic DNA was extracted according to Nercessian et al. (2005). DNA pellets were dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.5). DNA concentrations were measured photometrically and checked for purity (ratio of light absorption at 260–280 nm) using a nanoquant plate reader (Infinite M200 Pro, Tecan, Germany). 16S rRNA gene amplification was performed from genomic DNA extracts from days 10, 40, 50, 60, and 70, considering cultivable bacterial abundance information (heterotrophic bacteria and potential pathogenic Vibrio) and bacterial disease evidence (white feces disease), which occurred at previous rearing cycles between 50 and 65 days of rearing (personal communication with shrimp farm owners).
DNA sequences of the V3–V4 hypervariable region of the 16S rRNA gene were obtained from amplicon sequencing with the primer set S-D-Bact-0314-b-S-17 (5′-CCTACGGGNGGCWGCAG-3′)/S-D-Bact-0785-a-A-21 (5′-GACTACHVGGGTATCTAAKCC-3′; Klindworth et al., 2013). Sequencing at LGC genomics (Berlin) was done on an Illumina MiSeq using the V3 Chemistry (Illumina) in a 2 × 300 bp paired-end run. Demultiplexing, i.e., grouping of sequences by sample, and the removal of the primer sequences from the raw paired-end reads were performed by LGC genomics (Berlin, Germany). Further bioinformatic analysis steps were carried out at ZMT, Bremen, Germany, according to Hassenrück et al. (2016). Sequences were quality trimmed with a sliding window of 4 bases and a minimum average quality of 15 with trimmomatic v.033 (Bolger et al., 2014). Quality trimmed sequences were merged using PEAR v0.9.8 (Zhang J. et al., 2014). The swarming approach was used to cluster OTUs using swarm v2.1.1 (Mahé et al., 2014). For each OTU, one representative sequence (seed sequence) was taxonomically classified with SINA (SILVA Incremental Aligner) v1.2.11 using the SILVA rRNA project reference database (release 128) at a minimum alignment similarity and quality of 0.9 and a last common ancestor consensus of 0.7 (Pruesse et al., 2012). Unwanted lineages (such as archaea, chloroplasts, and mitochondria), as well as singletons and doubletons, i.e., OTUs occurring only once or twice in the data set, were removed. Rarefaction curves were calculated based on OTU richness and inverse Simpson index to assess the quality of sequence data sets. Samples with fewer than 500 sequences were excluded from the dataset for the analysis.
DNA sequence datasets were deposited in the European Nucleotide Archive (ENA) with the project accession number PRJEB26390, using the data brokerage service of the German Federation for Biological Data (GFBio, Diepenbroek et al., 2014), while biogeochemical data were archived in PANGAEA1.
Detection of Virulence Genes via PCR
Detection of toxin genes, i.e., transcriptional regulator (toxR), thermolabile haemolysin (tlh), thermostable direct haemolysin (tdh), Photorhabdus insect-related (pirA and pirB) was performed in a Mastercycler® (Eppendorf, Nexus gradient, Hamburg, Germany) using the set of primers described in Supplementary Table 2. As these genes are common genes in several representative of the genus Vibrio (i.e., V. cholera, V. vulnificus, V. parahaemolyticus, V. alginolyticus, and V. owensii), we designed specific primer pairs which only amplify DNA sequences belonging to V. parahaemolyticus as this bacterium causes most of bacterial diseases in shrimp (Soto-Rodriguez et al., 2015; Heenatigala and Fernando, 2016; Xiao et al., 2017). Ten samples, which had a high amplicon abundance of Vibrio, were chosen for PCR analysis (Supplementary Table 3). The PCR conditions were as follows: a reaction mixture consisted of 2 μL (20 ng μL-1) of template, 2 μL of 10× PCR buffer B containing 15 mM MgCl2, 0.5 μL of 25 mM MgCl2, 0.5 μL of 0.2 mM dNTPs, 1 μL of 0.1 mM forward and reverse primer, 0.1 μL of 5U μL-1 Taq polymerase (all reagents provided by Roboklon EURx, Berlin, Germany) and 12.9 μL sterile distillated water. The amplification conditions were as follows: pre-denaturation at 95°C for 3 min, followed by 40 amplification cycles consisting of denaturation at 95°C for 30 s, annealing at 60°C for 15 s, and extension at 72°C for 30 s, and a final elongation at 72°C for 5 min. Vibrio parahaemolyticus DSM 11058 (DSMZ, Braunschweig, Germany) was used as positive control for those toxin genes, while Vibrio vulnificus DSM 10143 (DSMZ, Braunschweig, Germany) served as negative control.
Data Analysis
To examine differences in environmental parameters and bacterial abundances between culturing intensity (intensive and semi-intensive systems), and among days, as well as the interaction between culturing system and sampling day, general linear mixed models (GLMM) were performed with shrimp pond as random factor. Data were log-transformed to achieve normal distribution prior to statistical testing. Tukey’s post hoc tests were applied in cases where there were significant differences among sampling days and/or an interaction between sampling day and shrimp farming system. Forward model selection based on the Akaike information criterion (AIC), considering collinearity of the variables and variance inflation values, was used to determine environmental parameters, which best explained observed counts of cultivable heterotrophic bacteria and potential pathogenic Vibrio. Total shrimp harvest was tested with one-way ANOVA and total bacterial cell numbers from DAPI counting of FL and PA bacterial communities were analyzed using MANOVA. As first-level post hoc test, the effect of significant predictor variable (here: sampling day) in MANOVA of FL and PA bacterial numbers was then tested with individual GLMMs per fraction. Furthermore, pairwise comparisons between sampling days were conducted as second-level post hoc test. Repeated measures correlation between FL and PA bacterial cell numbers was estimated using rmcorr R package (Bakdash and Marusich, 2018).
Principal component analysis (PCA) was conducted to examine the relationship among environmental parameters, as well as cultivable bacterial abundances, and to characterize shrimp ponds of the different farming systems over time. Bray–Curtis dissimilarity coefficients were calculated to investigate variation of OTU compositions in FL and PA communities among ponds, as well as within and between the systems. Patterns of BCC in both fractions was visualized by non-metric multidimensional scaling (NMDS) based on Bray-Curtis dissimilarities using the function metaMDS of the vegan R package (Oksanen, 2017). Analysis of similarity (ANOSIM) was performed to test for differences in community composition in free-living and particle-attached fraction between systems over the time.
Redundancy analysis (RDA) was performed to explore relationships between bacterial communities and environmental variables in FL and PA fractions, for OTUs occurring in at least 10 samples, and with a sequence proportion of at least 1% in one sample. Data sets were separated by size fractions, and then centered log-ratio-transformed using aldex.clr (Fernandes et al., 2014) before being tested. Forward model selection based on the AIC, followed by collinearity and variance inflation inspection, was used to select the environmental parameters best suited to explain patterns in BCC. The significance of the individual parameters of the RDA models was assessed using restricted permutation tests to account for the repeated measurements within ponds. For the PA fraction, data from day 60 was excluded due to a missing value to ensure equal numbers of observation from each pond.
Population turnover, which occurred in the FL and PA fractions, was estimated for certain interesting OTUs of the genera Salegentibacter, Psychrobacter, Halomonas, and Vibrio, using the fraction of read abundance times cells method (FRAxC) proposed by Kevorkian et al. (2018), where bacterial cell numbers in the FL and PA of each selected sampling day were multiplied with the relative sequence proportion of 16S rRNA genes. We assumed that the biases associated with sampling procedure, DNA extraction, amplification, and cell counts did not vary over time. FRAxC results were also used to estimate correlation between estimated Vibrio abundance and selected environmental parameters (SPM, salinity, pH, temperature, ammonium, phosphate, and nitrite).
All statistical analyses, as well as figure visualizations were performed in R (R version 3.4.2, R Core Team, 2017, using R Studio v.0.98.1056) with the packages vegan (Oksanen, 2017), nlme (Pinheiro et al., 2017), ALDEx2 (Fernandes et al., 2014), rmcorr (Bakdash and Marusich, 2018), and gplots (Warnes et al., 2016).
Results
Shrimp Culturing Conditions
Total shrimp harvest differed significantly between semi-intensive and intensive systems (one-way ANOVA, F1,4 = 111.43, p < 0.01). The intensive system showed a twofold higher production than semi-intensive system, which were 3,950 ± 284 kg and 1,990 ± 151 kg, for intensive and semi-intensive final harvest (mean ± SD, n = 3 per system). There was a high variability among the ponds within each system for turbidity, chlorophyll a, pH, salinity, DO, and inorganic nutrients during rearing time, which increased over time (Table 1). Physical parameters such as salinity, pH, DO, chlorophyll a, and SPM were significantly different between the two systems (GLMM, p < 0.05, Supplementary Table 4). Among sampling days, we detected differences in temperature, salinity, pH, turbidity, chlorophyll a, SPM, cultivable heterotrophic bacteria and potential pathogenic Vibrio (GLMM, p < 0.05, Supplementary Table 4). The interaction between day and farming system was also significant for temperature, salinity, and turbidity. However, there were no significant differences between semi-intensive and intensive systems for dissolved inorganic nutrients, temperature, and turbidity (GLMM, p < 0.05, Supplementary Table 4).
Table 1.
Physical parameters, in-organic nutrient concentrations, and cultivable heterotrophic bacteria abundances over 70 rearing days in the semi-intensive and intensive farming system.
| Day | 10 |
20 |
30 |
40 |
50 |
60 |
70 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Systema | S | T | S | T | S | T | S | T | S | T | S | T | S | T |
| Parametersb | ||||||||||||||
| SPM∗ (mg L-1) | 66.6 ± 8.0a |
68.7 ± 5.2a |
68.1 ± 4.9a |
70.3 ± 7.7a |
80.1 ± 12.2b |
91.5 ± 8.3b |
96.5 ± 5.7c |
122.0 ± 22.2c |
158.1 ± 4.0d |
202.6 ± 18.5d |
167.3 ± 5.5d |
187.6 ± 11.0d |
173.1 ± 2.2d |
171.3 ± 17.7d |
| Chl a∗ (mg L-1) | 13.34 ± 7.13ac |
5.47 ± 3.82a |
14.61 ± 1.49ae |
10.11 ± 5.04ab |
11.22 ± 0.96ab |
22.8 ± 19.8ae |
18.5 ± 5.5ae |
77.9 ± 32.1e |
20.7 ± 9.1ae |
63.5 ± 29.1ce |
30.9 ± 32.3ae |
33.7 ± 14.2bce |
16.9 ± 14.9acd |
68.1 ± 27.7de |
| pH∗ | 8.24 ± 0.04b |
8.18 ± 0.11b |
8.20 ± 0.14ab |
7.95 ± 0.07ab |
8.26 ± 0.22ab |
7.9 ± 0.1ab |
8.5 ± 0.1b |
8.0 ± 0.1b |
8.3 ± 0.2ab |
7.6 ± 0.5ab |
7.9 ± 0.1a |
7.8 ± 0.1a |
8.4 ± 0.5b |
8.1 ± 0.1b |
| DO∗ (mg L-1) | 6.13 ± 0.14 |
6.32 ± 0.1 |
6.4 ± 0.02 |
6.36 ± 0.13 |
5.90 ± 0.65 |
6.2 ± 0.1 |
6.1 ± 0.2 |
6.9 ± 0.8 |
5.9 ± 0.3 |
6.1 ± 0.2 |
5.7 ± 0.1 |
6.3 ± 0.2 |
5.8 ± 0.5 |
6.2 ± 0.3 |
| Salinity∗ (PSU) | 35.37 ± 0.15bd |
38.43 ± 0.78d |
35.20 ± 0.69ad |
36.79 ± 0.50cd |
32.44 ± 3.26a |
35.9 ± 0.9bd |
33.7 ± 0.7abc |
35.0 ± 0.9abc |
33.4 ± 0.3ab |
35.8 ± 0.3bd |
33.6 ± 1.4abc |
34.1 ± 0.8abc |
34.6 ± 0.9abc |
33.6 ± 0.5ab |
| Temperature (°C) | 31.24 ± 0.41cd |
30.19 ± 0.50bcd |
28.08 ± 0.64a |
30.44 ± 0.18bcd |
31.61 ± 0.98d |
30.4 ± 1.6bcd |
29.6 ± 0.2ac |
28.9 ± 0.5ab |
31.0 ± 0.1cd |
30.1 ± 0.4bcd |
30.8 ± 0.8cd |
30.2 ± 0.1bcd |
30.7 ± 0.5bcd |
30.5 ± 0.4bcd |
| Turbidity (NTU) | 16.53 ± 10.69ac |
3.9 ± 2.21a |
24.5 ± 8.67bc |
9.93 ± 8.44ab |
35.87 ± 30.26bc |
22.9 ± 14.6bc |
27.0 ± 10.5bc |
25.7 ± 2.7bc |
25.9 ± 12.9bc |
25.3 ± 5.6bc |
27.6 ± 7.2bc |
30.2 ± 14.4c |
26.9 ± 9.2bc |
35.7 ± 15.9c |
| Cultivable Bact. (log10 CFU mL-1) | ||||||||||||||
| THB | 4.75 ± 0.06ac |
4.42 ± 0.15a |
5.35 ± 0.04bcd |
4.65 ± 0.30ab |
6.29 ± 0.15ef |
5.50 ± 0.16cd |
5.99 ± 0.50de |
6.76 ± 0.20eg |
6.87 ± 0.69fg |
7.44 ± 0.19g |
7.53 ± 0.19g |
7.41 ± 0.36g |
7.47 ± 0.18g |
7.4 ± 0.1g |
| TPPV | 2.03 ± 0.56a |
2.13 ± 0.90a |
4.35 ± 0.36bc |
3.32 ± 0.32ab |
4.51 ± 0.19bc |
3.98 ± 0.18b |
3.85 ± 0.29b |
3.73 ± 0.41b |
3.85 ± 0.48b |
3.72 ± 0.15b |
4.16 ± 0.69bc |
5.41 ± 0.89c |
4.18 ± 0.42bc |
4.36 ± 0.7bc |
| Nutrient (mg L-1) | ||||||||||||||
| NH4+ | 0.60 ± 0.37 |
0.436 ± 0.671 |
0.46 ± 0.71 |
0.15 ± 0.08 |
0.31 ± 0.23 |
0.26 ± 0.34 |
0.23 ± 0.19 |
0.26 ± 0.22 |
0.50 ± 0.07 |
0.37 ± 0.30 |
0.61 ± 0.858 |
0.835 ± 0.248 |
0.33 ± 0.34 |
0.36 ± 0.25 |
| NO2- | 0.004 ± 0.004 |
0.183 ± 0.292 |
0.004 ± 0.004 |
0.008 ± 0.007 |
0.021 ± 0.025 |
0.005 ± 0.003 |
0.007 ± 0.007 |
0.201 ± 0.334 |
0.016 ± 0.006 |
0.001 ± 0.001 |
0.002 ± 0.001 |
0.079 ± 0.113 |
0.013 ± 0.009 |
0.004 ± 0.003 |
| NO3- | 0.048 ± 0.06 |
0.075 ± 0.11 |
0.007 ± 0.005 |
0.097 ± 0.16 |
0.004 ± 0.003 |
0.027 ± 0.042 |
0.028 ± 0.02 |
0.213 ± 0.184 |
0.034 ± 0.025 |
0.014 ± 0.013 |
0.052 ± 0.04 |
0.307 ± 0.456 |
0.028 ± 0.026 |
0.284 ± 0.484 |
| PO43- | 0.33 ± 0.31 |
0.53 ± 0.41 |
0.29 ± 0.20 |
0.50 ± 0.37 |
0.22 ± 0.17 |
0.22 ± 0.27 |
0.31 ± 0.38 |
0.72 ± 0.07 |
0.39 ± 0.26 |
0.11 ± 0.12 |
0.35 ± 0.36 |
0.59 ± 0.47 |
0.61 ± 0.08 |
0.31 ± 0.22 |
| SiO44- | 1.424 ± 1.453 |
0.206 ± 0.191 |
0.179 ± 0.047 |
0.315 ± 0.253 |
1.217 ± 0.918 |
0.343 ± 0.134 |
0.565 ± 0.576 |
0.324 ± 0.245 |
0.898 ± 1.193 |
0.45 ± 0.36 |
0.636 ± 0.792 |
0.11 ± 0.07 |
0.623 ± 0.668 |
1.157 ± 1.413 |
Significant differences between systems at a significance threshold of 0.05 are indicated by asterisks; superscript letters indicate significant differences for the interaction system and day (p < 0.05). Data are shown as mean ± standard deviation. aS, semi-intensive system; T, intensive system. bSPM, suspended particulate matter; Chl a, chlorophyll a; THB, total heterotrophic bacteria; TPPV, total potential pathogenic bacteria.
In both systems, pH decreased rapidly after day 40, but when limestones were added into the ponds, it increased from 7.92 ± 0.13 to 8.38 ± 0.53 and 7.63 ± 0.46 to 8.11 ± 0.13, in semi-intensive and in intensive system, respectively. Salinity decreased from 35.37 ± 0.15 to 32.44 ± 3.26 PSU and 38.43 ± 0.78 to 33.57 ± 0.45 PSU, in semi-intensive and in intensive system, respectively, due to freshwater input. Overall, the intensive system showed significantly higher salinities, SPM, and chlorophyll a concentration than the semi-intensive system, as well as higher variability of inorganic nutrients (Table 1).
The abundances of cultivable heterotrophic bacteria (THB) and potential pathogenic Vibrio (TPPV) in semi-intensive and intensive systems increased with rearing time, and peaked at day 60, at concentrations of 3.4 × 107 CFU mL-1 of THB and 1.4 × 104 CFU mL-1 of TPPV, and 2.6 × 107 CFU mL-1 of THB and 2.65 × 105 CFU mL-1 of TPPV, respectively (Table 1). Forward model selection showed that SPM, salinity, turbidity, temperature, and phosphate were major determinants for THB, while the determinants for TPPV were SPM, salinity, turbidity, pH, temperature, nitrite, and silicate.
Principal component analysis for environmental parameters and cultivable bacterial abundances was able to retrieve 42.61% of the variation among ponds on the first two principal components (Figure 1). According to water parameters and cultivable bacterial abundances, there was a clear separation by shrimp farming systems based on rearing time. At the beginning of rearing (days 10 and 20), ponds of intensive and semi-intensive systems were clustered with high values of the first principal component, which was driven mainly by high salinity. Afterward, they were separated by the second principal component with ponds of the intensive system characterized by high concentrations of chlorophyll a. The abundances of THB and TPPV highly correlated to SPM (Spearman correlation, rho: 0.87 and rho: 0.40 for THB and TPPV, respectively).
FIGURE 1.
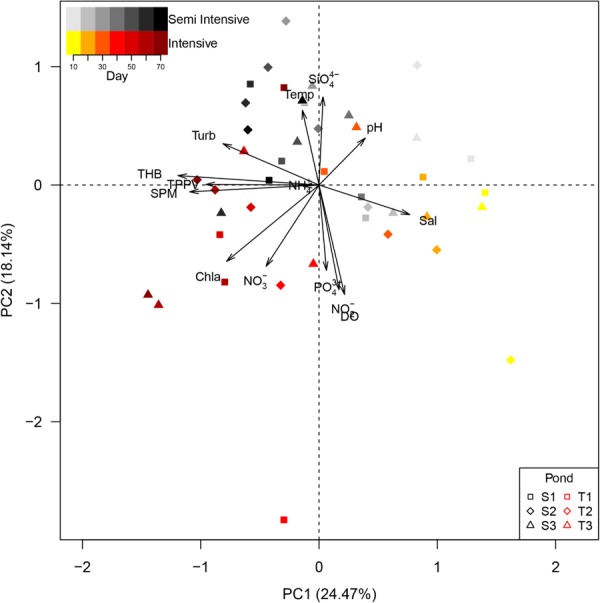
Principal Component Analysis of environmental parameters and bacterial abundances in intensive (T) and semi intensive (S) systems. Point shape indicates replicate pond of the same system. Increasing color intensity indicates rearing time. SPM, suspended particulate matter; THB, total heterotrophic bacteria; TPPV, total potential pathogenic Vibrio; NH4+, ammonium; Turb, turbidity; Temp, temperature; SiO44-, silicate; Sal, salinity; PO43+, phosphate; NO2-, nitrite; DO, dissolved oxygen; NO3-, nitrate; Chl a, chlorophyll a.
Aggregates containing bacterial cells were found in different sizes. At day 10, small aggregates appeared which were formed by bacterial cells and their exudates. After 40 days, aggregates were composed of plankton, bacterial cells and bacterial exudates, causing the increase of aggregate sizes in both systems, as well as bacterial cell numbers. Aggregates of sizes between 937.5 and 1,406.5 μm2 were most abundant in both system over time, with the highest aggregate number at day 10 (511 ± 63 aggregates mL-1) and at day 40 (494 ± 27 aggregates mL-1), containing 86 ± 3 and 72 ± 6 bacterial cells per individual aggregate for semi-intensive and intensive pond waters, respectively (mean ± SD; n = 3; Supplementary Figure 1 and Supplementary Table 1). Free-living (FL) bacterial cells of both systems increased and peaked at day 60 in concentration 3.8 × 107 cells mL-1 and 5.0 × 107 cells mL-1, for semi-intensive and intensive systems, respectively. Total particle-attached (PA) bacterial cell numbers were steady after day 40, with 6.8 × 105 cells mL-1 and 6.4 × 105 cells mL-1, for semi-intensive and intensive systems, respectively (Table 2). The FL cell numbers were positively correlated to the PA cell numbers (repeated measure correlation, Spearman correlation, rho = 0.50, df = 23, p = 0.01) and differed only among days (Supplementary Tables 5, 6).
Table 2.
Total bacterial cell numbers in the free-living (FL) and the particle attached (PA) fractions.
| Day | Fractions |
|||
|---|---|---|---|---|
| FL [×107 cells mL-1] |
PA [×105 cells mL-1] |
|||
| S | T | S | T | |
| 10 | 0.06 ± 0.01a | 0.09 ± 0.02a | 0.62 ± 0.08a | 0.50 ± 0.05a |
| 40 | 1.83 ± 0.13b | 3.09 ± 2.34b | 6.77 ± 0.50b | 6.40 ± 0.24b |
| 50 | 2.94 ± 1.04b | 2.13 ± 0.11b | 6.51 ± 0.79b | 6.55 ± 0.31b |
| 60 | 3.79 ± 0.89c | 5.01 ± 0.85c | 6.16 ± 0.67b | 6.09 ± 0.37b |
| 70 | 3.47 ± 0.94bc | 3.64 ± 1.22bc | 7.68 ± 0.36c | 7.42 ± 0.21c |
Data are shown as average ± standard deviation. Similar superscript letters after cell numbers in semi-intensive (S) and intensive (T) indicate no significant difference. Bacterial numbers in the FL and PA are interpreted separately. Complete statistical results were presented in Supplementary Table 6.
Bacterial Community Analysis
After removal of low-quality reads, 8,994–78,615 sequences, with on average 34,471 sequences per sample, were obtained from 59 samples. One sample was removed due to low sequence counts (<500 sequences). A total of 77,433 OTUs was obtained, ranging from 426 to 2465 OTUs, with an average of 1,312 OTUs per sample (Supplementary Figure 2).
Most abundant bacterial OTUs in both systems belonged to the classes Acidimicrobiia, Actinobacteria, Alphaproteobacteria, Bacilli, Cyanobacteria, Flavobacteriia, and Gammaproteobacteria (Figure 2). Among them, the genera Alteromonas, Erythrobacteraceae, Exiguobacterium, Halomonas, Vibrio, Pseudoalteromonas, Psychrobacter, Salegentibacter, and Sulfitobacter were present in every sample. Cyanobacteria, such as Synechococcus and Cobetia, were frequently present in the semi-intensive system. The highest sequence proportion in the FL fraction of both systems belonged to Salegentibacter, Sulfitobacter, and Halomonas, while in the PA fraction Psychrobacter, Vibrio, and Halomonas were dominant, where the latter comprised up to 80% of the sequences in samples from the semi-intensive system after 40 and 50 days (Figure 2). In the SILVA reference database (SILVA 128 version), Halomonas OTU1 has a sequence similarity of 98 and 100% to H. aquamarina and H. meridiana, respectively.
FIGURE 2.
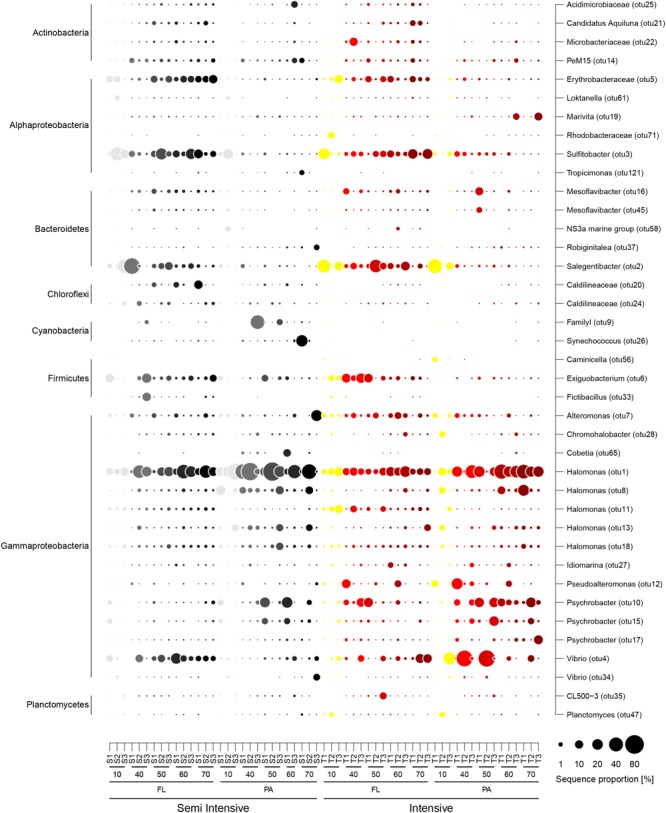
Contribution of the most abundant bacterial operational taxonomic units (OTUs) in the semi-intensive (light gray to black) and the intensive systems (yellow to dark red). Taxonomic affiliation for OTUs is provided for genus (Right) and class (Left) levels. S1–3 and T1–3: replicate pond for the semi-intensive and intensive ponds, respectively. Days of rearing are indicated below the pond symbols. FL, free-living fraction; PA, particle-attached fraction.
Free-living and particle-associated bacterial communities from the same water sample were very different from each other as indicated by Bray–Curtis dissimilarity coefficients (Table 3). In addition, there was high variability in BCC within the same fraction among replicate ponds, which lead to a wide range in Bray-Curtis dissimilarity coefficients (0.27-0.97 and 0.33-0.83 for PA and FL fractions, respectively) between intensive and semi-intensive systems (Table 4).
Table 3.
Bray–Curtis dissimilarities of bacterial community composition between free-living (FL) and particle-attached (PA) fractions.
| Pond | Day |
||||
|---|---|---|---|---|---|
| 10 | 40 | 50 | 60 | 70 | |
| S1 | 0.54 | 0.83 | 0.83 | 0.85 | 0.91 |
| S2 | 0.53 | 0.48 | 0.77 | NA∗ | 0.54 |
| S3 | 0.79 | 0.81 | 0.66 | 0.59 | 0.92 |
| Average | 0.62 | 0.71 | 0.75 | 0.72 | 0.79 |
| T1 | 0.45 | 0.51 | 0.57 | 0.74 | 0.91 |
| T2 | 0.68 | 0.87 | 0.91 | 0.49 | 0.69 |
| T3 | 0.63 | 0.68 | 0.70 | 0.58 | 0.82 |
| Average | 0.59 | 0.69 | 0.73 | 0.60 | 0.80 |
S, semi-intensive system; T, intensive; NA∗ indicated a missing sample for particle-attached fraction.
Table 4.
Bray–Curtis dissimilarities of bacterial community composition within system (among ponds) and between the systems for each fraction over the time.
| Fraction | Day | System | Within system |
Between system |
||
|---|---|---|---|---|---|---|
| Minimum | Maximum | Minimum | Maximum | |||
| FL | 10 | S | 0.68 | 0.73 | 0.33 | 0.83 |
| T | 0.66 | 0.85 | ||||
| 40 | S | 0.62 | 0.92 | 0.63 | 0.84 | |
| T | 0.47 | 0.69 | ||||
| 50 | S | 0.50 | 0.68 | 0.40 | 0.76 | |
| T | 0.58 | 0.83 | ||||
| 60 | S | 0.41 | 0.60 | 0.41 | 0.69 | |
| T | 0.46 | 0.64 | ||||
| 70 | S | 0.44 | 0.59 | 0.45 | 0.63 | |
| T | 0.55 | 0.66 | ||||
| PA | 10 | S | 0.69 | 0.71 | 0.70 | 0.98 |
| T | 0.78 | 0.96 | ||||
| 40 | S | 0.38 | 0.82 | 0.50 | 0.98 | |
| T | 0.52 | 0.91 | ||||
| 50 | S | 0.62 | 0.76 | 0.43 | 0.99 | |
| T | 0.50 | 0.95 | ||||
| 60 | S | 0.86 | 0.86 | 0.50 | 0.79 | |
| T | 0.50 | 0.65 | ||||
| 70 | S | 0.94 | 0.97 | 0.27 | 0.97 | |
| T | 0.62 | 0.65 | ||||
FL, free-living fraction; PA, particle-attached fraction; S, semi-intensive; T, intensive.
NMDS showed a highly heterogeneous composition of the bacterial communities in the FL and the PA fraction for both systems (Figure 3). Analysis of similarity (ANOSIM) confirmed that there was no detectable pattern in FL or PA the BCC between system and day (Supplementary Table 7).
FIGURE 3.
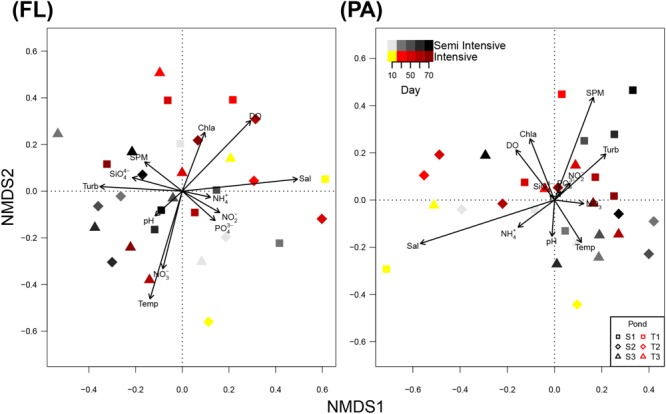
Non-metric multidimensional scaling (NMDS) plot of bacterial community composition (BCC) in the free-living (FL) and particle-attached (PA) fraction. Information on environmental parameters was added to the NMDS plot using envfit. Point shape indicates replicate ponds of the same system. Increasing color intensity indicates rearing time. SPM, suspended particulate matter; NH4+, ammonium; Turb, turbidity; Temp, temperature; SiO44-, silicate; Sal, salinity; PO43+, phosphate; NO2-, nitrite; DO, dissolved oxygen; NO3-, nitrate; Chl a, chlorophyll a.
Redundancy analysis revealed that environmental variables explained 20.53 and 36.77% of the BCC in the FL and the PA fractions, respectively (Supplementary Figure 3). Among the observed environmental parameters, salinity was best suited to explain patterns in the composition of both FL and PA bacterial communities (R2 > 10%). Furthermore, chlorophyll a and nitrate had a minor effect on BCC in the FL fractions (Table 5).
Table 5.
Contribution and significance of observe environmental factors which explain the variation in community composition of the free-living (FL) and the particle-attached (PA) fractions based on redundancy analysis (RDA).
| Source of variation | Adjusted R2 | df | F | P-value |
|---|---|---|---|---|
| FL | ||||
| Complete model (system + day) | 0.103 | 5, 24 | 1.669 | 0.005 |
| Sampling time (day) | 0.070 | 4, 24 | 1.543 | 0.007 |
| System | 0.042 | 1, 24 | 2.172 | 0.204 |
| Complete model (Sal + Chl a + NO3-) | 0.153 | 3, 26 | 2.749 | 0.001 |
| Salinity (Sal) | 0.112 | 1, 26 | 4.558 | 0.001 |
| Chlorophyll a (Chl a) | 0.047 | 1, 26 | 2.510 | 0.005 |
| Nitrate (NO3-) | 0.030 | 1, 26 | 1.944 | 0.017 |
| PA | ||||
| Complete model (system + day) | 0.035 | 4, 19 | 1.207 | 0.184 |
| Complete model (Sal) | 0.128 | 1, 22 | 4.379 | 0.001 |
Toxin Gene Assay, Vibrio Occurrence and Correlation to Environmental Parameters
All tested samples were negative for toxR, tlh, and tdh. These results indicate that no common pathogenic V. parahaemolyticus occurred in both systems, even though the given samples displayed high sequence proportions of Vibrio, specifically OTU4. The sequence of Vibrio OTU4 was identical to 16S gene sequences in the SILVA reference database belonging to V. mytili, V. diabolicus, and V. parahaemolyticus. The estimated abundance (FRAxC) of the dominant Vibrio OTU differed strikingly from that of the other dominant OTUs, such as Halomonas, Psychrobacter, and Salegentibacter, suggesting an inverse relationship. When FRAxCs of Vibrio were overly high, the proportions of the other genera were low. In the PA fraction, FRAxCs of Vibrio were negatively correlated to FRAxCs Halomonas (Figure 4).
FIGURE 4.
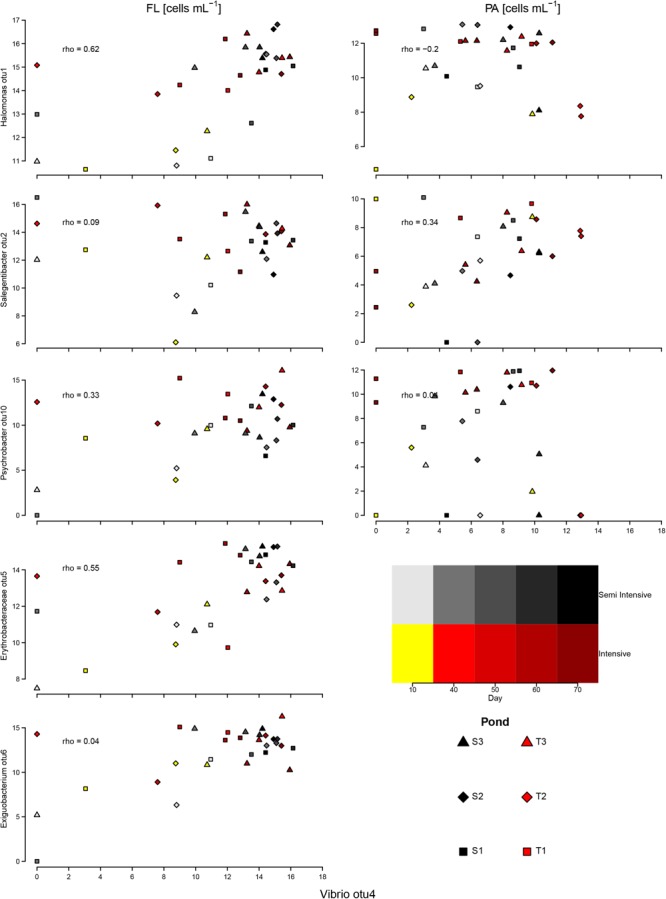
Correlation between estimated proportion of cells of the most-abundant OTUs in the FL and PA fractions and the dominant Vibrio OTU. The axes show log-transformed estimated bacterial numbers. rho, Spearman correlation coefficient.
FRAxCs of Vibrio in both fractions were positively correlated to SPM, temperature, ammonium, NP ratio, and TPPV, but they were negatively correlated to pH and salinity. In addition, Vibrio was negatively correlated to inorganic nitrite and phosphate in the FL fraction, although in the PA fraction the opposite trend was observed. In general, all correlation coefficients did not suggest strong correlation with values between -0.36 and 0.38 (Supplementary Table 8).
Discussion
No water discharge shrimp aquaculture resulted in an accumulation of organic matter, through feed pellets, shrimp feces and other organic materials. Previous studies reported that degradation of organic matter, leaching of feed pellets, and release of ammonium from feces increase inorganic nutrients in the water column (Funge-Smith and Briggs, 1998; Smith et al., 2002; Burford and Lorenzen, 2004). These nutrients support bacterial and phytoplankton growth, which then leads to simultaneous change of physical parameters such as pH, DO, SPM, and turbidity (Burford and Williams, 2001; Burford et al., 2003).
In this study, shrimp pond water parameters and respective BCC in intensive and semi-intensive systems were compared. Contrary to our expectations, higher shrimp density in intensive farming systems did not significantly increase inorganic nutrients in the water column. We suggest that excessive nutrients were rapidly taken up by bacteria and phytoplankton as their abundance increased more strongly in the intensive system. This was also reflected in high SPM values in the intensive system due to the aggregation of phytoplankton cells. Phytoplankton can take up total ammonia nitrogen (TAN), while heterotrophic bacteria perform nitrification and nitrogen assimilation (Avnimelech, 2014). In pond water, photosynthesis by phytoplankton and cyanobacteria increases DO, while respiration and microbial activities, such as nitrification and sulfur oxidation decrease pH (Boyd and Tucker, 2014). The higher number of phytoplankton (which was indicated by Chl a) in intensive ponds might increase DO concentration. We supposed that photosynthesis might increase oxygen concentration. However, we are aware that our DO measurements only constitute daylight conditions and that due to the higher organic matter loading of the intensive system and in the absence of light, oxygen depletion via respiration processes may lower DO concentration below the levels of the semi-intensive system. Even though the intensive system resulted in higher abundances of phytoplankton and cultivable heterotrophic bacteria, we could show that the concentrations of harmful inorganic nutrients, such as ammonium and nitrite, DO, and pH were still far from lethal values for shrimps, as mentioned in other studies (Schuler, 2008; Avnimelech, 2014; Furtado et al., 2015).
We observed fluctuation of particle (aggregate or flocs) number over rearing time. We supposed that lower pellet input, sinking or consumption of flocs might cause the fluctuation of particle numbers. Water movement and exudate secreted either by bacteria or plankton lead to particle agglomeration (Grossart et al., 2006; Gärdes et al., 2011), generating bigger aggregates/flocs. When the aggregates sink to the bottom, particle numbers might decrease, resulting low turbidity/clearer water. Unfortunately, we did not measure daily feed input, sinking rate of aggregates and particle consumption by shrimp.
Environmental factors shape the structure and function of microbial communities (Allison and Martiny, 2008). Previous studies have shown that the succession of microbial communities was influenced by combinations of chlorophyll a, total nitrogen (TN), PO43-, C/N ratio (Xiong et al., 2014), total phosphate, chemical oxygen demand (Zhang D. et al., 2014), and feed sources added into ponds (Qin et al., 2016). Of the parameters measured in this study, salinity was the most determinant variable which shaped BCC in the FL and particle-attached PA fractions, which concurred with previous studies (Yang et al., 2016; Hou et al., 2017; Kirchmann et al., 2017). We observed salinity fluctuation in both systems and supposed that rainfall and addition of sea water from reservoir pond to maintain rearing pond water level (∼140–150 cm) decreased salinity. During our study, rainfall could increase 10–20 cm of water level, while sea water was added more frequent in intensive ponds due to regular mud removal. However, it should be considered that the influences of salinity on the BCC in the FL fraction do not completely exclude the influence of other water parameters, such as unmeasured sulfur compound in water column like hydrogen sulfide (H2S) or sulfite which may explain the continually presence of Sulfitobacter in FL fraction.
Bacterial communities in intensive and semi-intensive systems were dominated by heterotrophic halophilic bacterial genera Alteromonas, Erythrobacter, Halomonas, Pseudoalteromonas, Salegentibacter, Sulfitobacter, and Psychrobacter. Halomonas and Psychrobacter, the salt-tolerant heterotrophic nitrifying bacteria (Chankaew et al., 2017), can oxidize ammonia in high concentration under saline conditions (Sangnoi et al., 2016). Sulfitobacter can oxidize sulfur and degrade high molecular weight dissolved organic matter (Bourne et al., 2004; Sosa et al., 2017). Other bacterial genera, such as Salegentibacter, Exiguobacterium, and Erythrobacter, were associated with shrimp shell degradation after molting (Sombatjinda et al., 2011). In our study, BCC differed from those reported by Sombatjinda et al. (2011) who analyzed L. vannamei earthen pond water with salinities close to freshwater. They identified Nitrosomonas, Flavobacterium, Exiguobacterium, Synechococcus, Burkholderia, Nitrosospira, and Nitrobacter as dominant bacteria (Sombatjinda et al., 2011). We assume that salinity caused the differences in BCC observed in our study, considering the salinity range of 32–38 PSU compared to 2–10 PSU in previous study (Sombatjinda et al., 2011). This is supported by a study of Chankaew et al. (2016), where common nitrifying bacteria, including Nitrosomonas, Nitrosococcus, Nitrosolobus, Nitrospira, Nitrococcus, and Nitrobacter, were absent in shrimp pond water with 20 PSU. Furthermore, the regular addition of commercial probiotics (personal communication with pond owners), which contained Bacillus sp., Pseudomonas sp, Nitrosomonas sp., Aerobacter sp., Nitrobacter sp., and Nitrosococcus sp. did not affect BCC in the FL and PA fractions of both systems, indicating that nitrification processes were generated by other microbes. We propose that in our study the dominance of particular heterotrophic halophilic bacteria may result in higher uptake of inorganic nutrient such as ammonium and nitrite (Sangnoi et al., 2016). Therefore, even in the intensive pond waters inorganic nutrients remained low. In our study, BCC changed over time, following a “resilience scenario” (Allison and Martiny, 2008), in which a replacement of bacterial taxa occurred due to environmental change, followed by a quick return to its pre-disturbance composition. This phenomenon can be clearly seen in the particle-attached fraction of the intensive system. When pH decreased below 8 at day 40 and 50, Vibrio replaced Halomonas as the most abundant OTU, but the sequence proportions of Halomonas recovered, after the pH increased at day 60 and 70. In contrast, BCC in the FL and the PA fractions of the semi-intensive system during the same period was similar while experiencing stable pH (above 8). We propose that pH was a disturbing factor for heterotrophic halophilic bacteria in our shrimp systems and lead to the change of BCC only in the intensive system. This is supported by Krause et al. (2012) who reported that slight changes in pH caused compositional shifts in marine bacterial communities.
Vibrio, a potential opportunistic pathogen for L. vannamei (Heenatigala and Fernando, 2016; Rungrassamee et al., 2016), has been found in higher proportions in the PA fractions of the intensive system. This might indicate that the particulate fraction, specifically marine aggregates, can accumulate potentially pathogenic bacteria, as suggested by other studies (Lyons et al., 2005; Froelich et al., 2013). Our hypothesis that the intensive system seemed to be more vulnerable to Vibrio outbreaks is supported by high abundance and recurrent presence of Vibrio spp. only in the intensive system. Therefore, it is necessary to maintain SPM and aggregate abundance and to avoid massive Vibrio growth due to the fact that Vibrio can convert from non-virulent to virulent under certain cell density threshold or if dramatic environmental changes occur (Zhou et al., 2012). Nevertheless, toxin genes (toxR, tlh, and tdh) of V. parahaemolyticus (Makino et al., 2003), as well as pirAB toxins for AHPND (Xiao et al., 2017) were not present in this study. We argue that other halophilic, potentially probiotic, bacteria might inhibit chromosome II replication, where those genes are located (Makino et al., 2003), as proposed by Defoirdt et al. (2011). As no other known probiotic taxa were detected in this study, we hypothesize that the presence of Halomonas in both systems might inhibit Vibrio. Recently, Halomonas aquamarina has been applied as probiotic in L. vannamei culture to oxidize ammonium and to prevent Vibrio growth, which then leads to an increase of survival rates of L. vannamei (Zhang et al., 2009; Suantika et al., 2013; Sangnoi et al., 2016). However, other unobserved microorganisms, such as bacteriophage, Bdellovibrio, Saccharomyces, Streptococcus, Streptomyces, and protists might also suppress Vibrio (Munro et al., 2003; Alagappan et al., 2010; Chavez-Dozal et al., 2013; Lakshmi et al., 2013; Kongrueng et al., 2017). Considering final shrimp harvests, the intensive system is the more economically promising system. However, shrimp pond management needs be improved to maintain water quality as well as beneficial bacterial communities. Regular feed control and mud discard were likely to have affected BCC in our study. Without regular mud discard, sludge and organic matter degradation might increase oxygen demand, and as a result oxygen might become depleted in the bottom of the shrimp pond. Under these conditions, anaerobic degradation of organic matter produces H2S, and increases other toxic inorganic compounds such as ammonia and nitrite. Furthermore, sludge deposit in bottom pond might enlarge the anaerobic area which may reduce the habitable space for the shrimp. While strictly adjusted feeding in semi-intensive system seemed to lead to a more stable BCC, regular mud discard in the intensive system, which was conducted once to twice per day, might have prevented the accumulation of toxic compounds (i.e., hydrogen sulfide, ammonium, and nitrite), as indicated by the lower proportion of Sulfitobacter. We suggest that giving adequate feed, discarding sludge, maintaining pH, and salinity, might avoid abrupt water quality change, eliminate large anaerobic areas at the pond bottom, and may minimize proliferation of potential pathogens, especially Vibrio. Some aspects of shrimp pond management related studies are given in Hopkins et al. (1993); Funge-Smith and Briggs (1998), and Avnimelech and Ritvo (2003). Consequently, it is necessary for pond aquaculture to provide sufficient seawater and sludge reservoirs to avoid environmental degradation risks due to sludge removal.
In view of BCC in FL and PA fraction over rearing period and the absence of virulence genes, we propose to apply “bioflocs” in shrimp aquaculture. This can be generated by adding carbohydrates such as molasses, rice bran or tapioca. Bioflocs might become an alternative food source for shrimps and be able to maintain ammonia and nitrite concentration in shrimp pond (Avnimelech, 2014). Moreover, because bacteria from commercial probiotics were not detected in any samples, we suggest that the regular application of these probiotics in such high salinity rearing pond is not necessary to be done. At the end, operational shrimp rearing costs, especially for feed pellet and commercial probiotics, can be reduced.
Conclusion
Different stocking densities influenced water quality parameters, especially SPM, DO, chlorophyll a, and pH. High salinity shaped the BCC in the PA and FL fraction, favoring heterotrophic halophilic bacteria which facilitate an optimum uptake of inorganic nutrients and prevent increase of Vibrio growth. Despite a wide variability of BCCs in intensive and semi-intensive systems, the abundance of Vibrio was higher in the intensive system, which may therefore be more vulnerable to disease outbreaks. Monitoring BCC, especially the PA fraction’s, and larger particulates such as aggregates in shrimp pond waters may potentially prevent disease outbreaks such as Vibriosis, white feces disease, white-tale disease, and AHPND.
Ethic Statement
This study did not sacrifice any organism, nor introduce viable pathogenic bacteria to the environment. All sampling procedures were already informed to and have been agreed upon by shrimp pond owners.
Author Contributions
YA, JH, and AG designed the project. YA collected the samples and conducted the in situ measurement with the logistic support of AT and AK. YA and CH completed the statistical analysis. YA prepared the manuscript with input from all co-authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Mr. Edi Sujiyanto (owner of small and medium enterprise Sokowati), Mr. Andi Fatosa and Mr. Iwan Thomasfa (owner of small and medium enterprise Mandalika Agung), where we collected our samples and used their facilities during research.
Funding. We acknowledge financial support from the Leibniz Association. YA gets a funding for pursuing his Ph.D. with the RISET-Pro scholarship from Indonesian Ministry of Research, Technology and Higher Education.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02457/full#supplementary-material
References
- Alagappan K. M., Deivasigamani B., Somasundaram S. T., Kumaran S. (2010). Occurrence of Vibrio parahaemolyticus and its specific phages from shrimp ponds in East Coast of India. Curr. Microbiol. 61 235–240. 10.1007/s00284-010-9599-0 [DOI] [PubMed] [Google Scholar]
- Allison S. D., Martiny J. B. H. (2008). Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U.S.A. 105 11512–11519. 10.1073/pnas.0801925105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. L., Valderrama D., Jory D. (2016). “Shrimp production review,” in Global Aquaculture Alliance: Presentation Global Aquaculture Production Data and Analysis (Guangzhou: GOAL; ) 1–50. [Google Scholar]
- Avnimelech Y. (2014). Biofloc Technology – A Practical Guide Book 3rd Edn. Baton Rouge, LA: The World Aquaculture Society; 258 10.13140/2.1.4575.0402 [DOI] [Google Scholar]
- Avnimelech Y., Kochva M., Diab S. (1994). Development of controlled intensive aquaculture systems with limited water exchange and adjusted carbon to nitrogen ratio. Isr. J. Aquac. 46 119–131. [Google Scholar]
- Avnimelech Y., Ritvo G. (2003). Shrimp and fish pond soils: processes and management. Aquaculture 220 549–567. 10.1016/S0044-8486(02)00641-5 [DOI] [Google Scholar]
- Bakdash J. Z., Marusich L. R. (2018). rmcorr: Repeated Measures Correlation. R Package Version 0.3.0. Available at: https://CRAN.R-project.org/package=rmcorr [Google Scholar]
- Bidle K., Fletcher M. (1995). Comparison of free-living and particle-associated bacterial communities in the Chesapeake Bay by stable low-molecular-weight RNA analysis. Appl. Environ. Microbiol. 61 944–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone L. (1931). Anomuran, macruran Crustacea from Panama and Canal Zone. Bull. Am. Mus. Nat. Hist. 63 137–189. [Google Scholar]
- Bourne D. G., Young N., Webster N., Payne M., Salmon M., Demel S., et al. (2004). Microbial community dynamics in a larval aquaculture system of the tropical rock lobster, Panulirus ornatus. Aquaculture 242 31–51. 10.1016/j.aquaculture.2004.08.047 [DOI] [Google Scholar]
- Boyd C. E., Tucker C. S. (2014). “pH,” in Handbook for Aquaculture Water Quality ed. Burns J. (Auburn, AL: Craftmaster Printers; ) 95–112. [Google Scholar]
- Brun E., Rodgers C., Georgiadis M., Bjørndal T. (2009). “Economic impact of disease and biosecurity measures,” in Proceedings of International Biosecurity Conference; 2009 August 17-18 (Trondheim, Norwey: IABC; ) 1–34. [Google Scholar]
- Burford M. A., Lorenzen K. (2004). Modeling nitrogen dynamics in intensive shrimp ponds: the role of sediment remineralization. Aquaculture 229 129–145. 10.1016/S0044-8486(03)00358-2 [DOI] [Google Scholar]
- Burford M. A., Thompson P. J., McIntosh R. P., Bauman R. H., Pearson D. C. (2003). Nutrient and microbial dynamics in high-intensity, zero-exchange shrimp ponds in Belize. Aquaculture 219 393–411. [Google Scholar]
- Burford M. A., Williams K. C. (2001). The fate of nitrogenous waste from shrimp feeding. Aquaculture 198 79–93. [Google Scholar]
- Chankaew S., O-thong S., Songnoi Y. (2017). Nitrogen removal efficiency of salt-tolerant heterotrophic nitrifying bacteria nitrogen removal efficiency of salt-tolerant heterotrophic nitrifying bacteria. Chiang Mai J. Sci. 44 1–10. [Google Scholar]
- Chankaew S., O-thong S., Songnoi Y. (2016). “Halomonas sp. SKNB4, a proficient ammonium oxidizing bacterium,” in Proceedings of the 3rd national meeting on biodiversity management in Thailand June 15-17 (Copenhagen: Fund for Sustainable Education (FUSE)) 86–191. [Google Scholar]
- Chavez-Dozal A., Gorman C., Erken M., Steinberg P. D., McDougald D., Nishiguchi M. K. (2013). Predation response of Vibrio fischeri biofilms to Bacterivorus protists. Appl. Environ. Microbiol. 79 553–558. 10.1128/AEM.02710-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzon G., Lawrence A., Gaxiola G., Rosas C., Guillaume J. (2004). Nutrition of Litopenaeus vannamei reared in tanks or in ponds. Aquaculture 235 513–551. 10.1016/j.aquaculture.2003.12.022 [DOI] [Google Scholar]
- De Schryver P., Defoirdt T., Sorgeloos P. (2014). Early mortality syndrome outbreaks: a microbial management issue in shrimp farming? PLoS Pathog. 10:e03919. 10.1371/journal.ppat.1003919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defoirdt T., Sorgeloos P., Bossier P. (2011). Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr. Opin. Microbiol. 14 251–258. 10.1016/j.mib.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Diepenbroek M., Glöckner F. O., Grobe P., Güntsch A., Huber R., König-Ries B., et al. (2014). “Towards an integrated biodiversity and ecological research data management and archiving platform: the German Federation for the curation of biological data (GFBio),” in Proceedings of the Informatik 2014 – Big Data Komplexität meistern. GI-Edition: Lecture Notes in Informatics (LNI). GI edn. Vol. 232 eds Plödereder E., Grunske L., Schneider E., Ull D. (Bonn: Köllen Verlag; ) 1711–1724. [Google Scholar]
- Fernandes A. D., Reid J. N., Macklaim J. M., McMurrough T. A., Edgell D. R., Gloor G. B. (2014). Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2:15. 10.1186/2049-2618-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich B., Ayrapetyan M., Oliver J. D. (2013). Integration of Vibrio vulnificus into marine aggregates and its subsequent uptake by Crassostrea virginica oysters. Appl. Environ. Microbiol. 79 1454–1458. 10.1128/AEM.03095-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funge-Smith S. J., Briggs M. R. P. (1998). Nutrient budgets in intensive shrimp ponds: implications for sustainability. Aquaculture 164 117–133. 10.1016/S0044-8486(98)00181-1 [DOI] [Google Scholar]
- Furtado P. S., Fugimura M. M. S., Monserrat J. M., Souza D. M., Garcia L., de O., et al. (2015). Acute effects of extreme pH and its influences on the survival and biochemical biomarkers of juvenile white shrimp, Litopenaeus vannamei. Mar. Freshw. Behav. Physiol. 48 417–429. 10.1080/10236244.2015.1086539 [DOI] [Google Scholar]
- Gärdes A., Iversen M. H., Grossart H. P., Passow U., Ullrich M. (2011). Diatom-associated bacteria are required for aggregation of Thalassiosira weissflogii. ISME J. 5 436–445. 10.1038/ismej.2010.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Aquaculture Alliance [GAA] (2013). Cause of EMS Shrimp Disease Identified. Available at: https://www.aquaculturealliance.org/blog/cause-of-ems-shrimp-disease-identified/ [accessed 05 March 2018]. [Google Scholar]
- Grossart H.-P., Czub G., Simon M. (2006). Algae-bacteria interactions and their effects on aggregation and organic matter flux in the sea. Environ. Microbiol. 8 1074–1084. 10.1111/j.1462-2920.2006.00999.x [DOI] [PubMed] [Google Scholar]
- Hargreaves J. A. (2013). Biofloc Production Systems for Aquaculture. Stoneville, MS: SRAC Publ; 1–12. Available at: https://srac.tamu.edu/ [Google Scholar]
- Hassenrück C., Fink A., Lichtschlag A., Tegetmeyer H. E., de Beer D., Ramette A. (2016). Quantification of the effects of ocean acidification on sediment microbial communities in the environment: the importance of ecosystem approaches. FEMS Microbiol. Ecol. 92:fiw027. 10.1093/femsec/fiw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heenatigala P. P. M., Fernando M. U. L. (2016). Occurance of bacteria species responsible for vibriosis in shrimp pond culture systems in Sri Lanka and assesment of the suitable control measures. Sri Lanka J. Aquat. Sci. 21 1–17. 10.4038/sljas.v21i1.7481 [DOI] [Google Scholar]
- Hopkins J. S., Hamilton R. D., Sandifer P. A., Browdy C. L., Stokes A. D. (1993). Effect of water exchange rate on production, water quality, effluent characteristics and nitrogen budgets of intensive shrimp ponds. J. World Aquacult. Soc. 24 304–320. [Google Scholar]
- Hou D., Huang Z., Zeng S., Liu J., Wei D., Deng X., et al. (2017). Environmental factors shape water microbial community structure and function in shrimp cultural enclosure ecosystems. Front. Microbiol. 8:2359. 10.3389/fmicb.2017.02359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Krishnan K. P. (2017). Differences in free-living and particle-associated bacterial communities and their spatial variation in Kongsfjorden. Arctic. J. Basic Microbiol. 57 827–838. 10.1002/jobm.201700216 [DOI] [PubMed] [Google Scholar]
- Kautsky N., Rönnbäck P., Tedengren M., Troell M. (2000). Ecosystem perspectives on management of disease in shrimp pond farming. Aquaculture 191 145–161. 10.1016/S0044-8486(00)00424-5 [DOI] [Google Scholar]
- Kementerian Kelautan dan Perikanan [KKP] (2015). Kelautan Dan Perikanan Dalam Angka Tahun 2015. Indonesia: Pusat Data, Statistik dan Informasi; 308. [Google Scholar]
- Kepner R. L., Pratt J. R. (1994). Use of fluorochromes for direct enumeration of total bacteria in environmental samples: past and present. Microbiol. Rev. 58 603–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevorkian R., Bird J. T., Shumaker A., Lloyd K. G. (2018). Estimating population turnover rates by relative quantification methods reveals microbial dynamics in marine sediment. Appl. Environ. Microbiol. 84:e01443-17. 10.1128/AEM.01443-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharisma A., Manan A. (2012). Kelimpahan bakteri Vibrio sp., pada air pembesaran udang vannamei (Litopenaeus vannamei) sebagai deteksi dini serangan penyakit vibriosis. J. Ilmiah Perikanan Kelautan 4 129–134. [Google Scholar]
- Kiørboe T., Tang K., Grossart H. -P., Ploug H. (2003). Dynamics of microbial communities on marine snow aggregates: colonization, growth, detachment, and grazing mortality of attached bacteria. Appl. Environ. Microbiol. 69 3036–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmann D. L, Cottrel M. T., DiTullio G. R. (2017). Shaping of bacterial community composition and diversity by phytoplankton and salinity in the Delaware Estuary, USA. Aquat. Microb. Ecol. 78 93–106. 10.3354/ame01805 [DOI] [Google Scholar]
- Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongrueng J., Mitraparp-Arthorn P., Bangpanwimon K., Robins W., Vuddhakul V., Mekalanos J. (2017). Isolation of Bdellovibrio and like organisms and potential to reduce acute hepatopancreatic necrosis disease caused by Vibrio parahaemolyticus. Dis. Aquat. Organ. 124 223–232. 10.3354/dao03120 [DOI] [PubMed] [Google Scholar]
- Kramer A. M., Lyons M. M., Dobbs F. C., Drake J. M. (2013). Bacterial colonization and extinction on marine aggregates: stochastic model of species presence and abundance. Ecol. Evol. 3 4300–4309. 10.1002/ece3.789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause E., Wichels A., Giménes L., Lunau M., Schilhabel M. B., Gerdts G. (2012). Small changes in pH have direct effects on marine bacterial community composition: a microcosm approach. PLoS One 7:e47035. 10.1371/journal.pone.0047035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmi B., Viswanath B., Gopal D. V. R. S. (2013). Probiotics as antiviral agents in shrimp aquaculture. J. Pathog. 2013 424123. 10.1155/2013/424123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavilla-Pitogo C. R., Leaño E. M., Paner M. G. (1998). Mortalities of pond-cultured juvenile shrimp, Penaeus monodon, associated with dominance of luminescent vibrios in the rearing environment. Aquaculture 164 337–349. 10.1016/S0044-8486(98)00198-7 [DOI] [Google Scholar]
- Lyons M. M., Ward J. E., Gaff H., Hicks R. E., Drake J. M., Dobbs F. C. (2010). Theory of island biogeography on a microscopic scale: organic aggregates as islands for aquatic pathogens. Aquat. Microb. Ecol. 60 1–13. 10.3354/ame01417 [DOI] [Google Scholar]
- Lyons M. M., Ward J. E., Smolowitz R., Uhlinger K. R., Gast R. J. (2005). Lyons, M. Maille, J. Evan Ward, Roxanna Smolowitz, Kevin R. Uhlinger, and Rebecca J. Gast. Lethal marine snow: pathogen of bivalve mollusc concealed in marine aggregates. Limnol. Oceanogr. 50 1983–1988. [Google Scholar]
- Mahé F., Rognes T. T., Quince C., de Vargas C., Dunthorn M. (2014). Swarm: robust and fast clustering method for amplicon-based studies. PeerJ. 2:e593. 10.7717/peerj.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino K., Oshima K., Kurokawa K., Yokoyama K., Uda T., Tagomori K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361 743–749. 10.1016/S0140-6736(03)12659-1 [DOI] [PubMed] [Google Scholar]
- Martin J.-L. M., Veran Y., Guelorget O., Pham D. (1998). Shrimp rearing: stocking density, growth, impact on sediment, waste output and their relationships studied through the nitrogen budget in rearing ponds. Aquaculture 164 135–149. [Google Scholar]
- Munro J., Oakey J., Bromage E., Owens L. (2003). Experimental bacteriophage-mediated virulence in strains of Vibrio harveyi. Dis. Aquat. Organ. 54 187–194. 10.3354/dao054187 [DOI] [PubMed] [Google Scholar]
- Nercessian O., Noyes E., Kalyuzhnaya M. G., Lidstrom M. E., Chistoserdova L. (2005). Bacterial populations active in metabolism of C1 compounds in the sediment of Lake Washington, a freshwater lake. Appl. Environ. Microbiol. 71 6885–6899. 10.1128/AEM.71.11.6885-6899.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J. (2017). Vegan: Ecological Diversity. Available at: https://cran.r-project.org/web/packages/vegan/vignettes/diversity-vegan.pdf [Google Scholar]
- Pinheiro J., Bates D., DebRoy S., Sarkar D., Eispack, Heisterkamp S., VanWilligen B. (2017). Package “Nlme.” Repository CRAN. Available at: https://CRAN.R-project.org/package=nlme [Google Scholar]
- Pruesse E., Peplies J., Glöckner F. O. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28 1823–1829. 10.1093/bioinformatics/bts252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Hou J., Deng M., Liu Q., Wu C., Ji Y., et al. (2016). Bacterial abundance and diversity in pond water supplied with different feeds. Sci. Rep. 6:35232. 10.1038/srep35232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2017). R: A Language and Environment for Statistical Computing. Available at: https://www.R-project.org/ [Google Scholar]
- Riemann L., Winding A. (2001). Community dynamics of free-living and particle-associated bacterial assemblages during a freshwater phytoplankton bloom. Microb. Ecol. 42 274–285. 10.1007/s00248-001-0018-8 [DOI] [PubMed] [Google Scholar]
- Robertson L., Lawence A., Castille F. (1993). The effect of feeding frequency and feeding time on growth of Penaeus vannamei (Boone). Aquacult. Fish. Manage. 24 1–6. [Google Scholar]
- Rungrassamee W., Klanchui A., Maibunkaew S., Karoonuthaisiri N. (2016). Bacterial dynamics in intestines of the black tiger shrimp and the Pacific white shrimp during Vibrio harveyi exposure. J. Invertebr. Pathol. 133 12–19. 10.1016/j.jip.2015.11.004 [DOI] [PubMed] [Google Scholar]
- Sahu B. C., Adhikari S., Dey L. (2013). Carbon, nitrogen and phosphorus budget in shrimp (Penaeus monodon) culture ponds in eastern India. Aquacult. Int. 21 453–466. 10.1007/s10499-012-9573-x [DOI] [Google Scholar]
- Sangnoi Y., Chankaew S., O-Thong S. (2016). Indigenous Halomonas spp., the potential nitrifying bacteria for saline ammonium waste water treatment. Pak. J. Biol. Sci. 20 52–58. 10.3923/pjbs.2017.52.58 [DOI] [PubMed] [Google Scholar]
- Schuler D. J. (2008). Acute toxicity of ammonia and nitrite to white shrimp (L. vannamei) at low salinities. J. World Aquacult. Soc. 41 438–446. [Google Scholar]
- Smith D. M., Burford M. A., Tabrett S. J., Irvin S. J., Ward L. (2002). The effect of feeding frequency on water quality and growth of the black tiger shrimp (Penaeus monodon). Aquaculture 207 125–136. 10.1016/S0044-8486(01)00757-8 [DOI] [Google Scholar]
- Sombatjinda S., Boonapatcharoen N., Ruengjitchatchawalya M., Wantawin C., Withyachumnarnkul B., Techkarnjanaruk S. (2011). Dynamics of microbial communities in an earthen shrimp pond during the shrimp growing period. Environ. Nat. Resour. Res. 1 171–180. 10.5539/enrr.v1n1p171 [DOI] [Google Scholar]
- Sosa O. A., Repeta D. J., Ferrón S., Bryant J. A., Mende D. R., Karl D. M., et al. (2017). Isolation and characterization of bacteria that degrade phosphonates in marine dissolved organic matter. Front. Microbiol. 8:1786. 10.3389/fmicb.2017.01786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Rodriguez S. A., Gomez-Gil B., Lozano-Olvera R., Betancourt-Lozano M., Morales-Covarrubias M. S. (2015). Field and experimental evidence of Vibrio parahaemolyticus as the causative agent of acute hepatopancreatic necrosis disease of cultured shrimp (Litopenaeus vannamei) in northwestern Mexico. Appl. Environ. Microbiol. 81 1689–1699. 10.1128/AEM.03610-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland J. D. H., Parsons T. R. (1972). A Practical Handbook of Seawater Analysis 2nd Edn. Ottawa: The Alger Press Ltd. [Google Scholar]
- Suantika G., Aditiawati P., Astuti D. I., Khotimah Z. F. (2013). The use of indigenous probiotic Halomonas aquamarina and Shewanella algae for white shrimp (Litopenaeus vannamei Boone) hatchery productivity in zero water discharge system. J. Aquac. Res. Dev. 4:5 10.4172/2155-9546.1000194 [DOI] [Google Scholar]
- Walker P. J., Winton J. R. (2010). Emerging viral diseases of fish and shrimp. Vet. Res. 41:51 10.1051/vetres/2010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes A. G. R., Bolker B., Bonebakker L., Huber W., Liaw A., Lumley T., et al. (2016). Package “Gplots.” Repository CRAN. Available at: https://CRAN.R-project.org/package=gplots [Google Scholar]
- Xiong J., Dai W., Li C. (2016). Advances, challenges, and directions in shrimp disease control: the guidelines from an ecological perspective. Appl. Microbiol. Biotechnol. 100 6947–6954. 10.1007/s00253-016-7679-1 [DOI] [PubMed] [Google Scholar]
- Xiao J., Liu L., Ke Y., Li X., Liu Y., Pan Y., et al. (2017). Shrimp AHPND-causing plasmids encoding the PirAB toxins as mediated by pirAB-Tn903 are prevalent in various Vibrio species. Sci. Rep. 7:42177. 10.1038/srep42177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., Zhu J., Wang K., Wang X., Ye X., Liu L., et al. (2014). The temporal scaling of bacterioplankton composition: high turnover and predictability during shrimp cultivation. Microb. Ecol. 67 256–264. 10.1007/s00248-013-0336-7 [DOI] [PubMed] [Google Scholar]
- Yang C., Wang Q., Simon P. N., Liu J., Liu L., Dai X., et al. (2017). Distinct network interactions in particle-associated and free-living bacterial communities during a Microcystis aeruginosa bloom in a plateau lake. Front. Microbiol. 8:1202. 10.3389/fmicb.2017.01202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Ma L., Jiang H., Wu G., Dong H. (2016). Salinity shapes microbial diversity and community structure in surface sediments of the Qinghai-Tibetan Lakes. Sci. Rep. 6:25078. 10.1038/srep25078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Wang X., Xiong J., Zhu J., Wang Y., Zhao Q., et al. (2014). Bacterioplankton assemblages as biological indicators of shrimp health status. Ecol. Indic. 38 218–224. 10.1016/j.ecolind.2013.11.002 [DOI] [Google Scholar]
- Zhang J., Kobert K., Flouri T., Stamatakis A. (2014). PEAR: a fast and accurate illumina paired-end reAd mergeR. Bioinformatics 30 614–620. 10.1093/bioinformatics/btt593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Mai K., Tan B., Ai Q., Qi C., Xu W., et al. (2009). Effects of dietary administration of probiotic Halomonas sp. B12 on the intestinal microflora, immunological parameters, and midgut histological structure of shrimp, Fenneropenaeus chinensis. J. World Aquac. Soc. 40 58–66. 10.1111/j.1749-7345.2008.00235.x [DOI] [Google Scholar]
- Zhou J., Fang W., Yang X., Zhou S., Hu L., Li X., et al. (2012). A nonluminescent and highly virulent Vibrio harveyi strain is associated with “bacterial white tail disease” of Litopenaeus vannamei shrimp. PLoS One 7:e29961. 10.1371/journal.pone.0029961 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


