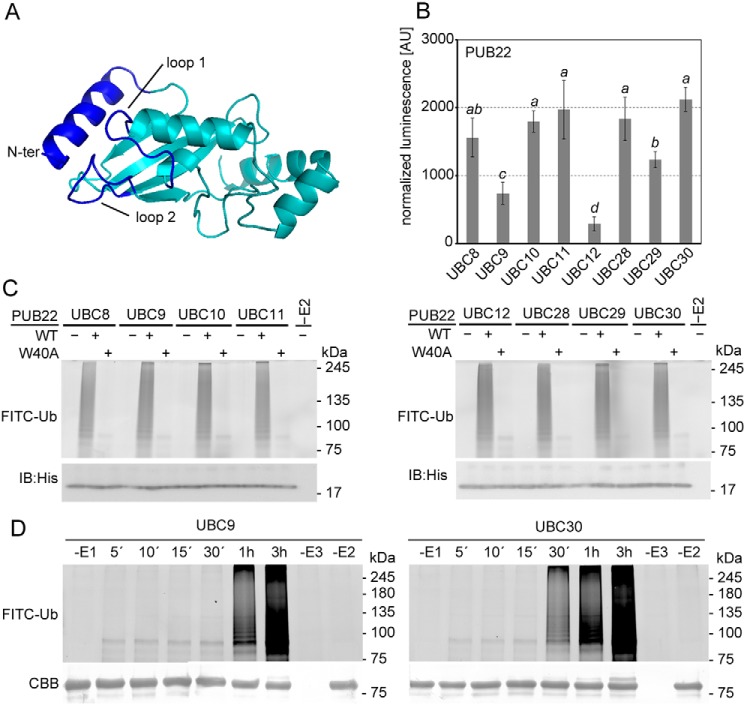
Figure 2.
PUB22 pairing selectivity with the highly homologous group VI E2s. A, structural model of UBC30 from group VI. U-box/RING interacting surfaces are highlighted in blue. B, SLCA of cLUC-PUB22 with nLUC-fused group VI E2s: UBC8, UBC9, UBC10, UBC11, UBC12, UBC28, UBC29, and UBC30, as indicated. Arabidopsis mesophyll protoplasts were transiently co-transformed with constructs containing the indicated genes. Transformation efficiencies were normalized by Renilla luciferase in the vector harboring the E2. Values indicate the average value of three independent biological experiments ± S.D. Statistically significant differences indicated by different letters were determined by one-way analysis of variance (ANOVA) and Tukey post hoc test (p < 0.05). C, in vitro autoubiquitination assay using MBP-tagged wildtype (WT) or W40A mutant variant of PUB22 (E3), His-UBA1 (E1), His-UBCs (E2s) from group VI, and fluorescein-tagged ubiquitin (FITC-Ub). D, in vitro time course autoubiquitination assay using MBP-PUB22, His-UBA1, His-UBC9, or His-UBC30, and FITC-Ub. Coomassie brilliant blue (CBB) shows equal loading. IB, immunoblot.