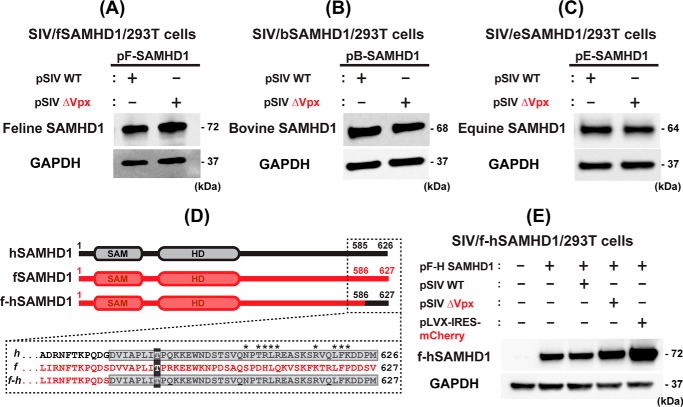
Figure 5.
Test for non–primate host SAMHD1 protein degradation by Vpx and fSAMHD1 C-T region for Vpx recognition. A–C, tests for Vpx-induced degradation of the non-primate host SAMHD1 proteins. 293T cells were transfected with pLVX-IRES-mCherry co-expressing HA-tagged SAMHD1 proteins from the three non-primate host species (A, fSAMHD1; B, bSAMHD1; and C, eSAMHD1) (0.1 μg) and either pSIV WT (2 μg) or pSIV ΔVpx (2 μg), and the SAMHD1 protein levels were determined by Western blotting with HA antibody. GAPDH was used as a loading control. D, construct of fSAMHD1 chimeric construct (f-hSAMHD1) containing the C-terminal Vpx recognition region of hSAMHD1. The sequence of the hSAMHD1 C-terminal region recognized by Vpx as well as the sequence of fSAMHD1 at the equivalent region is in dotted box. Asterisk sites are the positions of hSAMHD1 depicted for the Vpx recognition (PDB ID: 4CC9). The numbers indicate the amino acid positions of each SAMHD1. The f-hSAMHD1 chimera was constructed by swapping the fSAMHD1 C terminus (residues 586–627) with hSAMHD1 C terminus (residues 585–626) containing Vpx recognition region. The C-term phosphorylation sites (Thr-592 and Thr-593 in human and feline SAMHD1, respectively) are shaded in dark gray for reference. E, test for the degradation of f-hSAMHD1 chimeric construct by SIV. 293T cells were co-transfected with pLVX-IRES-mCherry co-expressing HA-tagged f-hSAMHD1 hybrid construct and mCherry protein (pF-H SAMHD1) (0.1 μg) and either pSIV WT (2 μg) or pSIV ΔVpx (2 μg). pLVX-IRES-mCherry expressing only mCherry protein (2 μg) was used as a negative control. The f-hSAMHD1 chimeric protein was detected by HA antibody. The Western blotting data presented in this figure are representative data from more than three independent transfections.