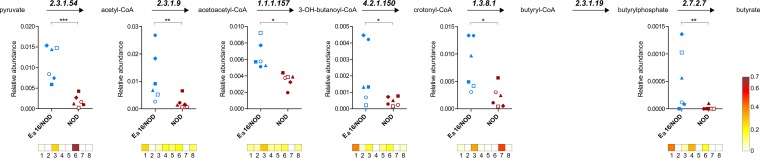
FIG 2.
Expression of butyrate biosynthesis enzymes in the fecal microbiotas of Eα16/NOD and NOD mice according to metaproteomic results. (Top) Schematic of butyrate biosynthetic pathway (from pyruvate to butyrate), with Enzyme Commission numbers corresponding to each enzyme. EC 2.3.1.54, formate acetyltransferase; EC 2.3.1.9, acetyl-CoA acetyltransferase; EC 1.1.1.157, 3-hydroxybutyryl-CoA dehydrogenase; EC 4.2.1.150, short-chain-enoyl-CoA hydratase; EC 1.3.8.1, butyryl-CoA dehydrogenase; EC 2.3.1.19, phosphate butyryltransferase; EC 2.7.2.7, butyrate kinase. (Middle) Scatterplots illustrating the relative abundance of each butyrogenic enzyme in the fecal microbiotas of Eα16/NOD and NOD mice; each dot represents a different mouse, with Eα16/NOD and NOD littermates reared in the same cage having the same shape. A paired sample test based on an inverted beta binomial model was used, with each pair of samples comprising two littermates with different genotypes. *, q value < 0.05; **, q value < 0.01; ***, q value < 0.001. (Bottom) Heatmaps showing the relative distribution of the taxonomic assignments (according to the lowest-common-ancestor principle) associated with each enzyme. Taxa associated with at least one enzyme and found in at least 4 mice are reported. Boxes: 1, Firmicutes; 2, Firmicutes, Clostridia; 3, Firmicutes, Clostridia, Clostridiales; 4, Firmicutes, Clostridia, Clostridiales, Clostridiaceae, Clostridium; 5, Firmicutes, Clostridia, Clostridiales, Eubacteriaceae, Eubacterium; 6, Firmicutes, Clostridia, Clostridiales, Lachnospiraceae; 7, Firmicutes, Clostridia, Clostridiales, Lachnospiraceae, Roseburia; 8, Firmicutes, Clostridia, Clostridiales, Oscillospiraceae, Oscillibacter.