High plasma protein binding (PPB) levels not only affect drug-target engagement but can also impact exposure of hepatocytes to antivirals and thereby affect antiviral activity. In this study, we assessed the effect of PPB on the antiviral activity of NVR 3-778, a sulfamoylbenzamide capsid assembly modulator (CAM).
KEYWORDS: HBV, capsid assembly modulators, plasma protein binding
ABSTRACT
High plasma protein binding (PPB) levels not only affect drug-target engagement but can also impact exposure of hepatocytes to antivirals and thereby affect antiviral activity. In this study, we assessed the effect of PPB on the antiviral activity of NVR 3-778, a sulfamoylbenzamide capsid assembly modulator (CAM). To this end, primary human hepatocyte (PHH) medium was spiked with plasma proteins. First, the effect of plasma proteins on the hepatitis B virus (HBV) infection assay was evaluated. The addition of plasma proteins neither decreased cell viability nor affected HBV DNA secretion or intracellular HBV RNA accumulation. In contrast, the secretion and intracellular amount of HBV proteins were induced with increasing amounts of plasma proteins. Next, the antiviral activity of NVR 3-778 was demonstrated by multiple assays while PPB and the time-dependent disappearance of the parent drug were quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Plasma proteins strongly decreased the free fraction of NVR 3-778, resulting in a physiologically relevant in vitro hepatocyte exposure. NVR 3-778 displayed a high PPB level, while the antiviral activity was reduced approximately only 4-fold. The disconnect between the high PPB level and the only moderate shift of the antiviral activity was explained by the rapid hepatic clearance of NVR 3-778 in the absence of plasma proteins. This study highlights the use of PHHs as a model to accurately determine the antiviral activity by capturing PPB, clearance, and liver distribution. It is advantageous to consider both pharmacokinetics and pharmacodynamics for selection of HBV antiviral drug candidates and for successful extrapolation of in vitro data to clinical studies.
INTRODUCTION
Hepatitis B virus (HBV) infection is a global public health problem, with more than 250 million chronic carriers that are at risk to develop liver disease and hepatocellular carcinoma (1). The viral particles contain a 3.2-kb partially double-stranded genome that encodes four major viral proteins (surface, core, polymerase, and X) from overlapping open reading frames (2). One of these proteins is the core protein that plays multiple roles in the viral life cycle of HBV, making it an attractive antiviral target. The core protein forms the nucleocapsid that is essential for encapsidation of the viral pregenomic RNA (pgRNA) and the viral polymerase. Within the nucleocapsid, pgRNA is reverse transcribed into the partially double-stranded relaxed circular DNA (rcDNA). Nucleocapsids are enveloped with lipid membrane containing HBV surface antigens (HBsAg) and secreted to infect neighboring hepatocytes. Alternatively, rcDNA containing nucleocapsids can be transported to the nucleus to maintain the covalently closed circular DNA (cccDNA) pool (3, 4). In addition, it has been suggested that core protein affects the epigenetic status of the nuclear cccDNA and is reported to interact with several host factors (5, 6).
New HBV antivirals are in development with the aim to achieve finite curative treatments. One promising group consists of capsid assembly modulators (CAMs) (7–11). CAMs interfere with the kinetics of core protein assembly and block encapsidation of the pgRNA. In addition, Berke et al. (8) and others demonstrated that CAMs, although at higher doses, can also prevent the formation of cccDNA in de novo-infected primary human hepatocyte (PHHs) (12, 13). At least two classes of CAMs have been described, those that lead to the formation of intact empty capsid structures devoid of HBV DNA (e.g., sulfamoylbenzamide derivates) and those that lead to aberrant core protein aggregates in the cell (e.g., heteroarylpyrimidines) (8, 9, 12, 14, 15).
NVR 3-778 (AL-3778) is a sulfamoylbenzamide CAM which has a demonstrated antiviral activity in a clinical study. In a phase 1b clinical trial, a mean serum HBV DNA reduction of 1.43 log10 IU/ml from baseline was demonstrated when NVR 3-778 was administered as monotherapy at a dose of 600 mg twice daily (BID) (16, 17; M. F. Yuen, E. J. Gane, D. J. Kim, F. Weilert, H. L. Y. Chan, J. Lalezari, S.G. Hwang, T. Nguyen, O. Flores, G. Hartman, S. Liaw, O. Lenz, T. N. Kakuda, W. Talloen, C. Schwabe, K. Klumpp, and N. Brown, unpublished data).
A key challenge for the development of antivirals is to predict the antiviral activity from in vitro studies to aid dose settings for in vivo clinical studies. Cell lines stably expressing HBV, such as HepG2.2.15 cells, are often used to predict the effective drug concentration required to inhibit in vitro HBV replication (18–20). Use of these cell lines remains limited, however, due to the poor expression of metabolic enzymes and alterations because of their tumorous origin (21, 22). Primary human hepatocytes (PHHs) have become a standard in characterizing novel anti-HBV agents since a complete HBV replication cycle can be studied (23–25). In addition, PHHs are the gold standard to evaluate hepatic drug uptake, drug distribution in liver, and metabolism, which are key parameters defining the pharmacokinetics of compounds, including HBV antivirals (26, 27).
The efficacy of a drug is highly dependent on its potency and the distribution from plasma to the target cells, specifically, in the case of HBV antivirals, hepatocytes. Plasma protein binding (PPB) is an important factor influencing the volume of distribution and drug clearance. According to the free-drug hypothesis, only the unbound drug is available for pharmacological action, and the efficacy will therefore be dependent on PPB (28, 29). The predominant plasma proteins are human serum albumin (HSA) and human alpha-1 acid glycoprotein (hA1GP). HSA is the most abundant protein present in human plasma, with concentrations ranging from 30 to 50 g/liter, while hA1GP is present at lower concentrations of approximately 0.7 g/liter. Moreover, both plasma proteins are produced by the liver itself (30, 31). For antivirals, drug concentrations substantially above the in vitro 90% effective concentration (EC90) are often necessary for potent inhibition of viral replication (32, 33). A good understanding of the effect of PPB on exposure and potency of drugs is necessary for reliable in vitro-to-in vivo extrapolations (IVIVEs) and clinical dose settings.
Here, we describe a new serum shift assay in PHHs cultured in plasma protein-relevant medium. NVR 3-778 was used as a representative compound with known high PPB levels from clinical trials to study and validate this assay. This assay provided an understanding of the impact of PPB on both the liver pharmacokinetics and pharmacodynamics of NVR 3-778. This physiologically relevant in vitro serum shift assay demonstrates the importance of PPB on liver distribution, clearance, and the efficacy of HBV antivirals and can be used to more accurately predict clinical exposures necessary for complete HBV inhibition.
RESULTS
Plasma protein binding and the free fraction of NVR 3-778 in protein-containing medium.
NVR 3-778 was highly bound to plasma proteins, with a free fraction between 1 to 2% in human plasma (Table 1). NVR 3-778 exhibited high binding (≥95.0% and <99.0%) to human serum albumin (HSA) and low binding (<50.0%) to human alpha-1-acid glycoprotein (hA1GP). The free fraction of NVR 3-778 in hepatocyte cultivation medium (HCM) containing 10% fetal calf serum (FCS) was approximately 73%, which results in a highly increased free-drug fraction compared to the physiologically relevant in vivo free-drug fraction (Table 1). The production of human albumin by in vitro-cultured PHHs was on average 0.005 g/liter (see Fig. S1A in the supplemental material). This amount of albumin secretion was considered negligible compared to the amount of HSA present in human plasma, which is approximately 1,000-fold higher (34). Binding of NVR 3-778 in HCM in the absence of additional plasma proteins was therefore believed to be solely to the plasma proteins present in fetal calf serum.
TABLE 1.
Free fraction of NVR 3-778 in different hepatocyte cultivation media and in human plasma
| Culture mediuma | Free fraction (%)b |
|---|---|
| no BSA, no hA1GP | 72.9 ± 1.9 |
| 4.3% BSA + 0.07% hA1GP | 9.7 ± 0.5 |
| 8% BSA + 0.07% hA1GP | 6.4 ± 0.1 |
| hPlasma | 1–2 |
Hepatocyte cultivation medium (HCM) was incubated with 1 μM NVR 3-778 while human plasma (hPlasma) was incubated with concentrations ranging from 0.1 to 10 μM.
Data represent mean values of three replicates ± SEM.
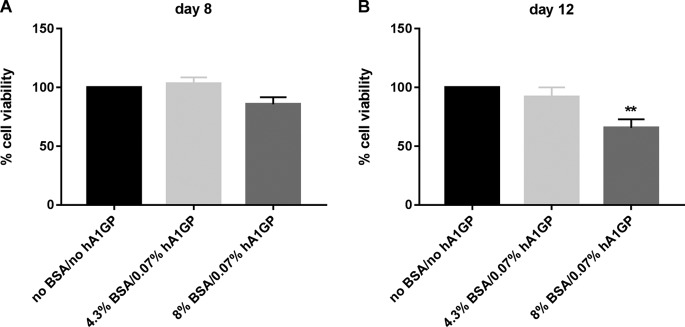
To evaluate the effect of physiologically relevant levels of human serum proteins on PHHs, HSA and hA1GP were added to hepatocyte cultivation medium (HCM) during the duration of PHH culturing (up to 8 days). For reasons that are unknown, cell viability was significantly decreased when PHHs were cultured in the presence of a physiological amount of HSA: 4.3% HSA reduced PHH viability by >50% after 8 days of cell culturing (Fig. S1B and C). Therefore, bovine serum albumin (BSA) was used as a substitute for HSA. HCM was spiked with 4.3% BSA and 0.07% hA1GP to mimic the protein composition of human plasma. Spiking of HCM with the physiological amount of plasma proteins decreased the free fraction of NVR 3-778 to 9.7%, which is higher than the free fraction found in human plasma, demonstrating species differences between human and bovine. To further decrease the free fraction, HCM was spiked with 8% BSA and 0.07% hA1GP, resulting in a free fraction of 6.4% and a more than 10-fold reduction of the free fraction compared to the free fraction measured in HCM without the addition of plasma proteins (Table 1). The effects of BSA and hA1GP on the cell viability of PHHs were assessed by measurement of the ATP content. Incubation of PHHs with HCM spiked with either 4.3% BSA and 0.07% hA1GP or with 8% BSA and 0.07% hA1GP for 8 days did not significantly reduce cell viability (Fig. 1A). Cell viability, however, was moderately and significantly affected when cells were cultured for an additional 4 days in HCM spiked with 8% BSA and 0.07% hA1GP but without any morphological changes (Fig. 1B and S2).
FIG 1.
Effect of plasma proteins on cell viability of HBV-infected PHHs. Cell viability was assessed by measurement of the ATP content after 8 and 12 days postinfection, as indicated. PHHs were HBV infected and cultured in medium without BSA/hA1GP or in medium spiked with either 4.3% BSA or 8% BSA together with 0.07% hA1GP for 8 and 12 days. Cell viability in HCM without BSA/hA1GP was set to be 100%. All data represent the means of three or more independent experiments ± SEM. **, P < 0.01.
Plasma proteins increase the susceptibility of PHHs to HBV.
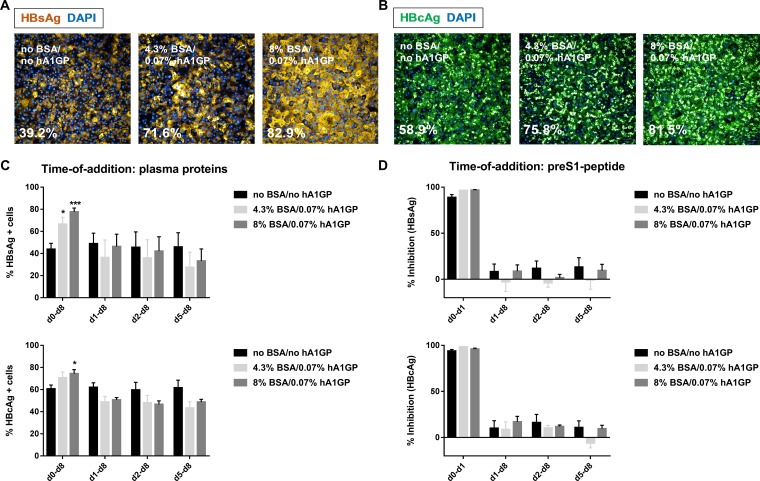
Initially, the impact of plasma proteins on HBV infectivity was assessed. The addition of plasma proteins to the viral inoculum (i.e., from day 0 onward) significantly induced the intracellular accumulation of HBsAg and HBV core antigen (HBcAg) (Fig. 2A and B and S3). For PHHs cultured in HCM without plasma proteins, the percentages of HBsAg- and HBcAg-positive cells were 39.2% and 58.9%, respectively. The percentages of HBsAg- and HBcAg-positive cells increased up to 80% in the presence of plasma proteins, yielding a homogenous population of HBV-infected cells.
FIG 2.
Effect of plasma proteins on the susceptibility of PHHs to HBV. Representative image of HBsAg-positive cells (A) and HBcAg-positive cells (B) in PHHs infected and cultured for 8 days in the absence and presence of plasma proteins. Scale bar, 50 μM. (C) Time of addition of plasma proteins. HBV-infected PHHs were incubated with plasma proteins from day (d) 0, day 1, day 2, or day 5 onward until day 8. The percentages of HBsAg- and HBcAg-positive cells were quantified by IF. (D) Time of addition of the pre-S1 peptide. HBV-infected PHHs were incubated with 0.5 μM pre-S1 peptide from day 0 until day 1 or from day 1, day 2, or day 5 onward until day 8. Percent inhibition was calculated based on the percentages of HBsAg- and HBcAg-positive cells in untreated HBV-infected PHHs cultured in the corresponding HCM. All data represent the means of three or more independent experiments ± SEM. *, P < 0.05; ***, P < 0.001.
Plasma proteins were added either together with the viral inoculum or at 1, 2, or 5 days postinfection to examine if plasma proteins improved the susceptibility of PHHs to HBV infection or increased viral replication. The number of HBV-infected PHHs was increased only when plasma proteins were added together with the viral inoculum at the time of HBV infection (Fig. 2C and S4A). In contrast, the percentages of HBsAg- and HBcAg-positive cells were comparable or even decreased when plasma proteins were added after the infection was established (Fig. 2C). These results suggest that plasma proteins enhance the susceptibility of PHHs to HBV infection rather than stimulate HBV replication. In addition, HBV-infected PHHs were treated with a pre-S1 peptide that blocks HBV entry to examine if plasma proteins induced reinfection. The pre-S1 peptide completely inhibited viral infection when it was added together with the viral inoculum in PHHs cultured in HCM both without and with plasma proteins (Fig. 2D and S4B). In contrast, pre-S1 peptide added to PHHs on day 1, day 2, or day 5 postinfection until the end of the infection experiment did not significantly reduce the number of HBsAg/HBcAg-positive cells in HCM compared to the level in untreated controls (Fig. 2D). These findings demonstrated that the increased number of HBV-infected cells when PHHs were cultured in HCM spiked with plasma proteins was an early event rather than one caused by reinfection at a later time point and that plasma proteins did not boost intracellular HBV replication.
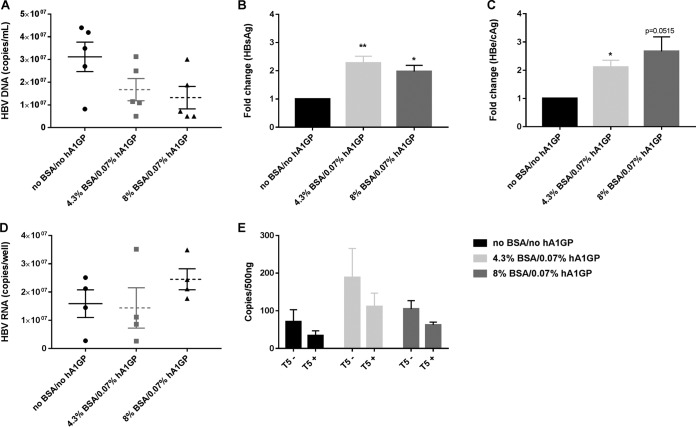
The release of HBV markers in the presence and absence of BSA and hA1GP in the culture medium was monitored on day 8 and day 12 postinfection. Secretion of HBV DNA in the culture medium was decreased by BSA and/or hA1GP (Fig. 3A and S5A). In contrast, HBsAg and HBV e antigen (HBeAg)/HBcAg secretion were both significantly increased compared to levels in HBV-infected PHHs cultured in HCM without plasma proteins (Fig. 3B and C and S5B and C). The increase of HBsAg and HBeAg/HBcAg secretion was less pronounced at day 12 than the increased secretion at day 8 postinfection. This might be related to the loss of infected cells due to increased cell toxicity when cells were cultured for an additional 4 days in plasma protein-containing medium (Fig. 1 and S5B and C). In addition, intracellular HBV RNA copy numbers were increased by the presence of plasma proteins (Fig. 3D). Although the detection of cccDNA remains challenging, quantitative PCR (qPCR) with cccDNA-specific primers demonstrated that cccDNA levels were moderately but not significantly increased when PHHs were infected with HBV in HCM spiked with plasma proteins compared to levels in PHHs infected with HBV in HCM without plasma proteins (Fig. 3E).
FIG 3.
Effect of plasma proteins on HBV DNA, HBsAg, and HBeAg/HBcAg in the supernatant of HBV-infected PHHs at 8 days postinfection. (A) HBV DNA secretion was analyzed by qPCR on DNA extracted from the cell culture supernatant of HBV-infected PHHs that were infected and cultured in HCM with or without the addition of plasma proteins. (B and C) An AlphaLisa assay was used to determine the amount of HBsAg and HBeAg/HBcAg that was secreted from HBV-infected PHHs that were infected and cultured in the absence and presence of plasma proteins. Results are expressed as fold change compared to levels in HBV-infected PHHs cultured in medium without BSA or hA1GP. (D) Total intracellular HBV RNA was extracted from HBV-infected PHHs that were infected and cultured in HCM with or without the addition of plasma proteins. HBV RNA was quantified by qPCR and normalized for β-actin. (E) cccDNA was quantified in nuclear DNA extracts by qPCR and normalized for the amount of nuclear DNA. Samples were digested with T5 exonuclease to digest contaminating rcDNA. T5−, nondigested; T5+; digested. All data represent the means of three or more independent experiments ± SEM.*, P < 0.05; **, P < 0.01.
Cytotoxicity and anti-HBV effect of NVR 3-778 in the presence of plasma proteins.
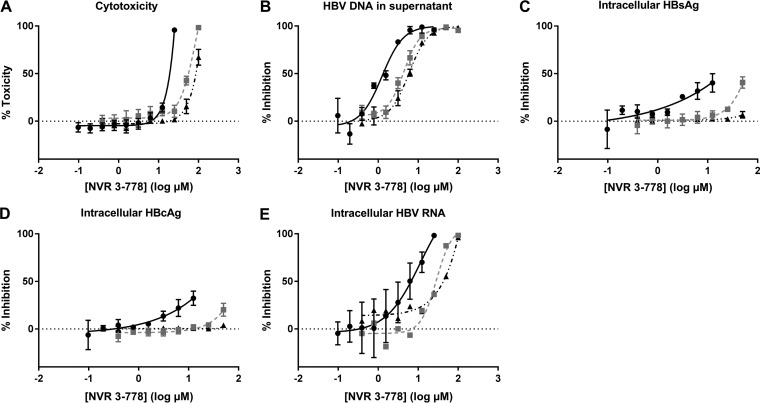
NVR 3-778 was either added together with the viral inoculum (i.e., from day 0) until day 8 or added from day 5 onward, when infection was already established, until day 12. Plasma proteins were added during virus inoculation (i.e., from day 0 onward) independent of the time of NVR 3-778 incubation. ATP content was used to determine cell toxicity induced by NVR 3-778. If NVR 3-778 was added from day 0 onward, the 50% cytotoxic concentration (CC50) value was 15.8 μM in the absence of plasma proteins, which was increased to 53.7 μM and 81.5 μM when PHHs were incubated in medium containing 4.3% BSA–0.07% hA1GP and 8% BSA–0.07% hA1GP, respectively (Table 2 and Fig. 4A). Cytotoxicity was reduced when PHHs were incubated with NVR 3-778 from day 5 onward. On day 12, no significant cytotoxicity was observed up to the highest concentration tested when NVR 3-778 was added to the HCM without plasma proteins (CC50 of >25 μM), while CC50 values were increased to 81.7 μM and 101.3 μM in plasma protein-containing medium (Table 2 and Fig. S6A). Differences in cytotoxicity levels between the times of addition of NVR 3-778 could be related to changes of metabolite formation due to long-term hepatocyte culturing (27, 35). The secretion of HBV DNA in the culture medium was dose-dependently inhibited by NVR 3-778 with similar 50% effective concentration (EC50) and EC90 values if NVR 3-778 was added together with the viral inoculum or when infection was established. The addition of 4.3% BSA–0.07% hA1GP resulted in a 2- to 3-fold reduction in antiviral potency, while spiking of HCM with 8% BSA–0.07% hA1GP resulted in a 4- to 5-fold reduction (Table 2 and Fig. 4B and S6B). Consistent with the previously reported effect of CAMs on blocking cccDNA formation during de novo infection, HBsAg and HBeAg/HBcAg secretion was inhibited only when NVR 3-778 was added together with the viral inoculum, thus confirming the effect of NVR 3-778 on the early events of the HBV viral life cycle (Fig. S6C to F). However, inhibition at the highest concentrations was related to cytotoxicity. In the presence of plasma proteins, inhibition of HBsAg and HBeAg/HBcAg secretion by NVR 3-778 was almost or even completely abolished. These results are in line with those obtained from monitoring intracellular HBsAg and HBcAg (Fig. 4C and D and S7). For immunofluorescence (IF) staining, cells were incubated with HBV inoculum and NVR 3-778 at concentrations ranging from 0 μM to 12.5 μM in HCM without plasma proteins and from 0 μM to 50 μM in HCM with plasma proteins. Since the highest concentrations used are lower than the CC50s under the two conditions, inhibition of HBV protein expression/staining is assumed not to be substantially affected by cytotoxicity. The percentages of both HBsAg- and HBcAg-positive cells were reduced, but not completely, in NVR 3-778-containing medium without plasma proteins and in HCM spiked with 4.3% BSA–0.07% hA1GP. In contrast, NVR 3-778 completely failed to block de novo HBV infection of PHHs when they were incubated in HCM spiked with 8% BSA–0.07% hA1GP (Fig. 4C and D and S7). Intracellular HBV RNA levels were dose-dependently inhibited if NVR 3-778 was added together with the viral inoculum at the time of infection. The anti-HBV activity of NVR 3-778 on the intracellular HBV RNA levels was serum shifted to an extent similar to that observed for the other HBV markers (Fig. 4E).
TABLE 2.
Cytotoxicity and anti-HBV effect of NVR 3-778 in HBV-infected PHHs in the presence and absence of plasma proteinsa
| Condition | Cytotoxicity (CC50 [μM])b |
EC50 for extracellular HBV DNA (μM)c |
EC90 for extracellular HBV DNA (μM)c |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Days 0–8 |
Days 5–12 |
Days 0–8 |
Days 5–12 |
Days 0–8 |
Days 5–12 |
|||||||
| Mean (95% CI) | Fold change | Mean (95% CI) | Fold change | Mean (95% CI) | Fold change | Mean (95% CI) | Fold change | Mean (95% CI) | Fold change | Mean (95% CI) | Fold change | |
| no BSA, no hA1GP | 15.85 (14.32–17.55) | >25 | 1.256 (0.792–1.945) | 0.906 (0.562–1.338) | 4.660 (2.356–15.740) | 4.267 (2.142–13.940) | ||||||
| 4.3% BSA + 0.07% hA1GP | 53.72 (48.92–58.99) | 3.4 | 81.68 (67.98–98.15) | >3 | 4.379 (3.567–5.406) | 3.5 | 2.468 (2.042–2.903) | 2.7 | 12.133 (8.236–20.324) | 2.6 | 9.451 (7.111–13.274) | 2.2 |
| 8% BSA + 0.07% hA1GP | 81.54 (74.25–89.54) | 5.1 | 101.30 (89.88–114.10) | >3 | 5.868 (4.831–7.066) | 4.7 | 3.712 (2.598–4.876) | 4.1 | 19.364 (13.335–32.137) | 4.2 | 20.137 (12.445–41.880) | 4.7 |
Data represent mean values of three or more independent experiments, and 95% confidence intervals (CI) are in parentheses.
Cytotoxicity was assessed by measurement of the ATP content. Fold change represents the change in the mean 50% cytotoxic concentration (CC50) in the presence of plasma proteins compared to the value in the absence of plasma proteins.
Extracellular HBV DNA was analyzed by extraction of DNA from the cell culture supernatant and quantified by qPCR. Fold change represents the value in the presence of plasma proteins compared to that in the absence of plasma proteins. EC50, 50% effective concentration; EC90, 90% effective concentration.
FIG 4.
Cell cytotoxicity and anti-HBV effect of NVR 3-778 on HBV DNA in the supernatant and on intracellular HBsAg, HBcAg, and HBV RNA. NVR 3-778 was added together with the viral inoculum until day 8. HBV-infected PHHs were incubated with a 2-fold dilution series of NVR 3-778 in HCM without plasma proteins (●) or in HCM spiked with either 4.3% BSA–0.07% hA1GP (■) or 8% BSA–0.07% hA1GP (▲). (A) Cell cytotoxicity was assessed by ATP content after incubation of HBV-infected PHHs with NVR 3-778. Percent toxicity was related to the ATP content of untreated PHHs cultured in the corresponding HCM. (B) HBV DNA production in the cell culture supernatant was quantified by qPCR. Percent inhibition was compared to level of HBV DNA produced by untreated cells cultured in the corresponding HCM. (C) Percentage of HBsAg-positive cells was quantified by IF. Percent inhibition was compared to the percentage of HBsAg-positive cells in untreated cells cultured in the corresponding HCM. (D) Percentage of HBcAg-positive cells was quantified by IF. Percent inhibition was compared to the percentage of HBcAg-positive cells in untreated cells cultured in the corresponding HCM. (E) Intracellular HBV RNA was quantified by qPCR. Percent inhibition was compared to the HBV RNA quantity in untreated cells cultured in the corresponding HCM. All data represent the means of three or more independent experiments ± SEM.
Effect of plasma protein binding on liver distribution and clearance of NVR 3-778.
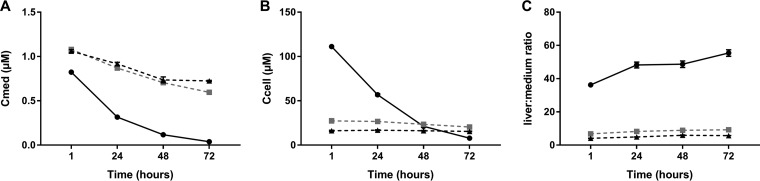
To study the pharmacokinetics of NVR 3-778 in the in vitro HBV infection assay, PHHs were incubated with 1 μM NVR 3-778 at day 5 postplating. The concentration of the parent drug was quantified in the hepatocyte lysates and in the culture medium after 1, 24, 48, and 72 h. NVR 3-778 was rapidly and completely cleared in culture medium without plasma proteins while the parent drug was only slowly cleared when PHHs were cultured in HCM spiked with plasma proteins (Table 3 and Fig. 5A). As a result, the apparent intrinsic in vitro hepatic clearance (CLint,app) levels were serum shifted 5.7- and 8.3-fold in HCM spiked with 4.3%–0.07% hA1GP and 8% BSA–0.07% hA1GP, respectively. According to the free-drug hypothesis, only the unbound drug is available for clearance, and the CLint,app value was corrected for the free fraction, resulting in a comparable in vitro unbound hepatic clearance (CLint,u) in all three HCM of approximately 150 μl/min/106 cells. Due to the extensive turnover of the parent drug in HCM without plasma proteins, the average exposure (average concentration, Cavg) of PHHs between day 5 and day 8 to NVR 3-778 was only 0.26 μM in HCM without plasma proteins. The Cavg was increased to 0.79 or 0.84 μM in HCM spiked with 4.3% BSA–0.07% hA1GP or 8% BSA–0.07% hA1GP, respectively (Table 3). In addition, the initial cell-associated concentration of NVR 3-778 was approximately 4- and 7-fold lower in PHHs cultured in HCM spiked with 4.3% BSA–0.07% hA1GP and 8% BSA–0.07% hA1GP, respectively. In HCM without plasma proteins, the cellular concentration rapidly decreased from 111.16 μM to 7.78 μM over time while in HCM spiked with plasma proteins, the cellular concentration was strongly reduced but remained stable due to delayed metabolism of NVR 3-778 (27.33 μM to 20.49 μM and 16.14 ± μM to 15.49 μM for PHHs cultured in HCM spiked with 4.3% BSA–0.07% hA1GP and 8% BSA–0.07% hA1GP, respectively) (Fig. 5B). As a result of the high PPB level and the decreased cellular concentrations, the cell-associated drug accumulation ratio (Kp) and mean predicted ratio of the concentration of drug in liver to that in medium were approximately 5.7 and 9.1-fold lower for PHHs cultured in HCM spiked with 4.3% BSA–0.07% A1GP and 8% BSA–0.07% A1GP, respectively, than in PHHs cultured in HCM without plasma proteins (Table 3 and Fig. 5C).
TABLE 3.
Pharmacokinetic properties of NVR 3-778 in PHHs cultured in HCM spiked with or without plasma proteins
| Condition | Mean value ± SEM (fold change)a |
|||||
|---|---|---|---|---|---|---|
| CLint,app (μl/min/106 cells) | CLint,u (μl/min/106 cells) | AUClast (ng · h/ml) | Cavg (μM) | Kp | Drug concn in liver/drug concn in mediumb | |
| no BSA, no hA1GP | 89.49 ± 5.33 | 122.75 ± 7.32 | 8,248 | 0.26 | 175.89 ± 14.90 | 47.12 ± 0.61 |
| 4.3% BSA + 0.07% hA1GP | 15.77 ± 0.45 (5.7) | 162.56 ± 4.61 (1.3) | 24,749 (3.0) | 0.79 (3.0) | 30.96 ± 2.02 (5.7) | 8.29 ± 0.20 (5.7) |
| 8% BSA + 0.07% hA1GP | 10.77 ± 0.14 (8.3) | 168.33 ± 2.22 (1.4) | 26,146 (3.2) | 0.84 (3.2) | 19.26 ± 1.58 (9.1) | 5.16 ± 0.05 (9.1) |
After 5 days of culturing, mock-infected PHHs were incubated with 1 μM NVR 3-778 for 4 days. Cells and supernatant were collected after 1, 24, 48, and 72 h to quantify turnover of the parent drug. Fold changes were calculated based on pharmacokinetic results obtained in PHHs cultured in HCM without BSA and hA1GP. CLint,app, intrinsic apparent clearance; CLint,u, unbound intrinsic clearance; AUClast, area under the concentration-time curve from time zero to the last measurable concentration; Cavg, average concentration; Kp, partition coefficient. Data represent mean values of three replicates ± SEM.
Ratio of the predicted concentration of NVR 3-778 per gram of liver tissue (nanograms/gram of liver tissue) to the concentration of NVR 3-778 in the medium (nanograms/milliliter), normalized for hepatocellularity.
FIG 5.
Pharmacokinetic properties of NVR 3-778 in PHHs cultured in different HCM. PHHs that were cultured for 5 days in either HCM without BSA/hA1GP (●) or in HCM spiked with 4.3% BSA–0.07% hA1GP (■) or with 8% BSA–0.07% hA1GP (▲) were incubated with 1 μM NVR 3-778 for 4 days, and both cells and supernatant were harvested at 1, 24, 48, and 72 h postincubation. The parent drug was quantified by LC/MS-MS at the indicated time points. (A) Quantification of the parent drug in the cell culture supernatant of PHHs. (B) Cellular concentration normalized for the hepatocyte volume of NVR 3-778 in PHHs. (C) Predicted liver-to-medium ratio of NVR 3-778 at the indicated time points calculated based on the medium and hepatocyte concentrations of NVR 3-778 in PHHs. Hepatocyte concentrations were normalized for the hepatocellularity to predict the corresponding liver concentrations. All data represent the means of three replicates ± SEM.
DISCUSSION
The hepatitis B virus (HBV) is an exclusively hepatotropic virus, and in vitro-cultured primary human hepatocytes (PHHs) are a very relevant culture model to assess HBV replication and inhibition thereof (23, 36). In addition, the liver is the main drug-metabolizing organ, and PHHs are the gold standard for in vitro hepatic clearance studies (37, 38). This dual role of PHHs was exploited in this study to understand the impact of PPB on both the pharmacokinetic and pharmacodynamic effects of NVR 3-778 to ultimately support the prediction of the human efficacious dose. Plasma protein binding (PPB) is a key parameter that determines the free-drug concentration in circulation and subsequently affects the pharmacokinetics, pharmacodynamics, and safety of compounds. Intrinsic hepatic clearance, drug transport, and distribution are all essential pharmacokinetic parameters that are influenced by PPB and that ultimately determine the exposure of a compound to the target organ (38–40). Many marketed drugs and compounds in discovery exhibit high PPB levels (>90%), and physiologically relevant preclinical assays are necessary to make reliable predictions on the impact of PPB (41). The effect of PPB on the in vitro compound activity can be studied by serum shift assays where cells are either cultured in the presence of dilutions of human serum up to 40% or incubated in medium in the presence of physiological concentrations of HSA and hA1GP (12, 42–46). The latter approach gives more reliable results than using human serum that contains confounding factors such as hormones and growth factors (47). In addition, culturing cells in 100% human serum is generally not possible due to cell viability issues. To the best of our knowledge, spiking PHH culture medium with plasma proteins to evaluate the effect of PPB on the activity of HBV antivirals has not been demonstrated. Using PHHs cultured in plasma protein-containing medium as an in vitro model, the effect of PPB on the antiviral activity, cellular exposure, and intrinsic clearance can be evaluated in the same system to more accurately predict clinical effective exposures.
Initially, the susceptibility and infection of PHHs with HBV cultured in the absence or presence of plasma proteins was evaluated. Culturing PHHs in medium spiked with HSA severely decreased cell viability (>50%) for reasons that are unknown. Therefore, BSA was used as a substitute for the human variant. Spiking of HCM with BSA and hA1GP did not decrease or only moderately decreased cell viability at 8 days postplating but significantly decreased PHH cell viability after 12 days of culturing, which could potentially be a limitation for long-term infection experiments. Surprisingly, the intracellular and secreted amounts of HBV proteins were highly and significantly increased when BSA and hA1GP were added at the time of HBV infection. The percentages of HBsAg- and HBcAg-positive cells increased from approximately 40% and 60%, respectively, to 80% when PHHs were infected in protein-containing medium. In addition, BSA and hA1GP induced the intracellular HBV RNA quantity and cccDNA content while the production of HBV DNA was decreased. It has previously been demonstrated that albumin is associated with HBV particles and that HBsAg bears an albumin receptor (48–50). The association between HBV particles and albumin might improve receptor binding and/or modify a posttranslational event, resulting in increased production of HBsAg and HBeAg/HBcAg. The increase in HBV proteins was lost when plasma proteins were added after the infection was established, indicating that BSA and hA1GP improved the susceptibility of PHHs to HBV rather than stimulating HBV replication. The decrease in viral DNA secretion is not understood but might be related to negative-feedback mechanism(s) because of the larger number of HBV-infected cells. The envelope protein of duck HBV was demonstrated to regulate cccDNA amplification through a negative-feedback mechanism; whether this also applies to human HBV remains unclear (51, 52). In addition, experiments with a pre-S1 peptide entry inhibitor demonstrated that BSA and hA1GP did not induce any reinfection, confirming that plasma proteins improve only the initial susceptibility of PHHs to HBV.
Due to species differences, binding of NVR 3-778 to bovine albumin was lower than that of human albumin. Nonetheless, the free fraction of NVR 3-778 was more than 7- and 10-fold reduced when PHHs were cultured in HCM spiked with 4.3% BSA–0.07% hA1GP and 8% BSA–0.07% hA1GP, respectively, approximating the free fraction measured in human plasma. The inhibition of the release of HBV DNA-containing particles in the supernatant, the primary mode of action of NVR 3-778, was only 3- to 5-fold reduced. The apparent discrepancy between the 10-fold-decreased free fraction and the only 5-fold-reduced antiviral activity was related to the hepatic drug clearance of NVR 3-778. In the absence of plasma proteins, NVR 3-778 was rapidly metabolized, resulting in decreasing hepatocyte exposures over the course of the experiments. Due to the high PPB level of NVR 3-778 in HCM spiked with plasma proteins, the rate of metabolism was decreased, resulting in lower but constant NVR 3-778 hepatocyte concentrations over the 4-day incubation period. The delayed metabolism due to a reduction of the free fraction can be beneficial for rapidly cleared compounds such as NVR 3-778, reducing the actual loss of antiviral potency. The reduction in hepatocyte exposure, however, was greater than the delay in metabolism and still resulted in a 4- to 5-fold reduction of the antiviral efficacy. In addition, active transport was not impacted by the high degree of PPB as NVR 3-778 is not a hepatic transporter substrate (data not shown). PPB would therefore mainly impact passive NVR 3-778 transport and metabolic clearance.
It is noteworthy that the reported EC50 of NVR 3-778 for inhibition of HBV DNA replication in the HepG2.2.15 cell line is approximately 0.47 μM and that the antiviral EC50 was increased 12-fold in the presence of 40% human serum (53; Yuen et al., unpublished data). This 12-fold serum shift is in accordance with the high degree of PPB of NVR 3-778 but is substantially overestimated compared to the actual serum shift observed in PHHs cultured in protein-relevant medium (only 5-fold). Hepatoma cell lines such as HepG2.2.15 cells express only low levels of cytochrome-metabolizing enzymes, resulting in higher average exposures to NVR 3-778 than with PHHs due to the lack of metabolic intrinsic clearance (21). This emphasizes that both PPB and hepatic intrinsic clearance are essential to predict the impact of PPB on the antiviral activity of HBV antivirals.
There are several potential advantages of CAMs compared to nucleos(t)ide analogues. In contrast to nucleos(t)ide analogues, CAMs act on multiple mechanisms for abrogating persistent HBV infection, such as inhibition of pgRNA encapsidation and prevention of cccDNA replenishment via intracellular reshuttling of the nucleocapsids (4, 7, 8). An additional advantage of CAMs is their ability to block de novo infection of PHHs, albeit at higher concentrations (8, 12, 13). NVR 3-778 inhibited, although not completely, de novo infection of PHH at higher concentrations in the absence of plasma proteins but failed to protect PHHs from de novo HBV infection at concentrations below the CC50 in the presence of plasma proteins. More potent and metabolically stable CAMs are needed to achieve exposures that are well tolerated and high enough to efficiently interfere with both HBV capsid assembly and de novo cccDNA formation.
The antiviral activity of NVR 3-778 was evaluated in a phase 1b clinical trial (16, 17; Yuen et al., unpublished data). The data from this study was retrospectively reanalyzed using the in vitro data obtained in PHHs in the presence of plasma proteins. NVR 3-778 dosing below 400 mg once a day (QD) for 28 days resulted in no or very limited HBV DNA changes from baselines in patients with trough concentration (Ctrough) values below the in vitro EC50 and EC90 values determined in HBV-infected PHHs cultured in medium spiked with 8% BSA–0.07% hA1GP. Dosing of 400 mg QD resulted in a mean (standard deviation [SD]) change of serum HBV DNA from baseline of −0.49 (0.67) log10 IU/ml. Consistent with the HBV DNA decline, mean (SD) Ctrough values of 3.7 (2.0) μM observed in this dosing group correspond to 1× the in vitro-predicted EC50 (mean EC50 of 3.7 μM in HCM spiked with 8% BSA–0.07% hA1GP). Moreover, Ctrough levels at 400 mg QD were 5.5-fold lower than the in vitro-predicted EC90 in plasma protein-relevant medium (mean EC90 of 20.1 μM in HCM spiked with 8% BSA–0.07% hA1GP) (16, 17; Yuen et al., unpublished data). Using EC50/EC90 data from PHHs cultured in the presence of plasma proteins could have predicted that higher doses were required to achieve serum HBV DNA declines larger than 0.5 log. Dosing of 600 mg BID for 28 days resulted in an in vivo mean (SD) Ctrough exposure of 19.4 (8.6) μM, which corresponds to 1× the in vitro-predicted EC90 (i.e., in HCM spiked with 8% BSA–0.07% hA1GP). This high exposure resulted in a mean 1.43-log10 IU/ml decline of serum HBV DNA from baseline over 28 days. The efficacy of NVR 3-778 was also evaluated in chimeric mice with humanized livers (54). This study demonstrated that twice-daily dosing of 405 mg/kg resulted in a decline of serum HBV DNA of 1.9 log10 IU/ml and Ctrough levels of 31 μM after 14 days of treatment. Ctrough values correspond to 1.5× the in vitro-predicted EC90 in plasma protein-relevant medium and are comparable to the Ctrough values and subsequent serum HBV DNA declines obtained in humans by dosing 600 mg BID.
As PHHs are both the target and drug clearance organ, they can be used for the parallel study of the pharmacokinetics and pharmacodynamics of newly developed HBV direct-acting antivirals. In vitro (serum shift) assays using PHHs are a valuable model for the development and study of HBV antivirals, including nucleoside analogues and especially those that display a high degree of PPB (>90%). For compounds with low PPB levels, hepatocyte exposure will mainly rely on hepatic uptake and hepatic intrinsic clearance, among other pharmacokinetic parameters. This novel in vitro serum shift assay was able to predict the efficacious dosage of NVR 3-778 for the treatment of chronic HBV carriers. This study emphasizes the importance of pharmacokinetics as well as pharmacodynamics to predict the exposures that need to be achieved to completely block HBV replication and potentially result in functional cure of HBV.
MATERIALS AND METHODS
Compounds.
NVR 3-778 and the pre-S1 peptide were synthesized in-house.
PHH infection assay.
Plateable, cryopreserved primary human hepatocytes (PHHs) (Hu1903 [Life Technologies] or HH1053 [In Vitro Admet Laboratories (IVAL)]) were thawed in universal cryopreservation recovery medium (UCRM) thawing medium (IVAL), centrifuged for 10 min at 100 × g, and plated at a density of 80,000 cells/well onto collagen I-coated 96-well plates (Corning) in 150 μl of UPCM universal plating medium (IVAL). The day after, PHHs were overnight inoculated with 2,500 genome equivalents (GE) per cell of cell culture-derived HBV (genotype D) in PHH cultivation medium (Dulbecco's modified Eagle medium [DMEM]) containing 10% fetal bovine serum (FBS) and 2% dimethyl sulfoxide (DMSO) supplemented with HEPES, l-proline, insulin, epidermal growth factor, dexamethasone, and ascorbic acid-2-phosphate (purchased from Life Technologies and Sigma) without additional plasma proteins (this medium is referred to as HCM without plasma proteins) or in hepatocyte cultivation medium (HCM) spiked with either 4.3% (43 g/liter) bovine serum albumin (BSA) (Sigma) and 0.07% (0.7 g/liter) human alpha-1 acid glycoprotein (hA1GP) (Sigma) or with 8% BSA (80 g/liter) and 0.07% hA1GP. For inoculation, PHH medium was supplemented with 4% polyethylene glycol (PEG) 8000 (Sigma). The day after infection, cells were extensively washed with DMEM, and medium was replaced with 150 μl of HCM either alone or spiked with plasma proteins. Thereafter, medium was changed every other 3 to 4 days. Compound was either added from day 0 together with the viral inoculum until day 8 or from day 5 until day 12. Compounds were diluted in DMSO, resulting in a 2% solution, and nine 2-fold dilutions were prepared from 0 to 25 μM for NVR 3-778 in HCM without addition of plasma proteins and from 0 to100 μM in HCM spiked with plasma proteins.
Cell viability assay.
ATPLite 1 step (PerkinElmer) was used to assess cell viability according to the manufacturer's protocol. Luminescence was measured using a ViewLux (PerkinElmer). The percent toxicity (TOX) was determined relative to the untreated control in the corresponding HCM, and 50% cytotoxic concentration (CC50) values were calculated with a four-parameter logistic (4PL) curve model using GraphPad Prism.
Quantification of viral HBV DNA in the cell culture supernatant.
On day 8 and day 12, viral DNA was extracted using a MagNAPure 96 DNA and viral NA small-volume kit (Roche). The HBV DNA copy number was quantified by qPCR with a standard curve from known quantities of HBV DNA. Primers 5′-GTGTCTGCGGCGTTTTATCA-3′ (sense) and 5′-GACAAACGGGCAACATACCT-3′ (antisense) in combination with an HBV probe, 5′-FAM-CCTCTKCATCCTGCTGCTATGCCTCATC-3′-TAMRA (where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine), were used for qPCR. Dose-response curves and 50% and 90% effective concentration (EC50 and EC90, respectively) values were calculated with a four-parameter logistic (4PL) curve model using GraphPad Prism.
Quantification of secreted HBeAg/HBcAg and HBsAg in the cell culture supernatant.
Secreted HBeAg/HBcAg and HBsAg were quantified as recently described by an amplified luminescent proximity homogenous assay developed in-house (AlphaLisa; PerkinElmer) (8). The amounts of HBeAg/HBcAg and HBsAg in the culture medium were quantified by using standard curves of recombinant HBsAg (developed in-house) or HBeAg (Jena Bioscience). Different standard curves were used for either plasma protein-free or protein-spiked HCM.
HBcAg and HBsAg immunofluorescence staining.
PHHs were fixed with 4% formaldehyde for 15 min at room temperature (RT). After fixation, cells were washed and permeabilized with 1% Triton X-100 in phosphate-buffered saline (PBS) for 10 min at RT and blocked overnight with 3% goat serum in PBS at 4°C. The next day, cells were incubated with the primary antibodies for 1 h at RT. An in-house anti-HBsAg antibody and a 1:1,600 dilution of a polyclonal rabbit anti-HBcAg (Dako) were used. Thereafter, cells were washed three times and incubated with a 1:500 dilution of the appropriate secondary antibody (Invitrogen) and Hoechst (Invitrogen) for 1 h at RT in PBS containing 3% goat serum. The cells were washed three times and stored in PBS. The plates were analyzed by high-content imaging (Opera Phenix; PerkinElmer) (magnification, ×20), and the percentage of infected cells was quantified by automated image analysis (Phaedra).
Quantification of intracellular HBV RNA.
Intracellular HBV RNA was extracted with an RNeasy 96 kit (Qiagen). Briefly, cells were lysed in 200 μl of RLT buffer containing 1% β-mercaptoethanol, and RNA was extracted according to the manufacturer's protocol. Upon RNA extraction, cDNA was synthesized by SuperScript III reverse transcriptase according to the kit instructions (ThermoScientific). For quantification of HBV RNA, the same primers and probe as described to quantify the amount of HBV DNA were used. β-Actin was used for normalization and amplified with the primers 5′-GGCCAGGTCATCACCATT-3′ (sense) and 5′-ATGTCCACGTCACACTTCATG-3′ (antisense) and probe 5′-Cy5-TTCCGCTGCTAOCCTGAGGCTCTC-3′-IAbRQSp. The β-actin copy number was quantified by qPCR with a standard curve from known quantities of β-actin.
cccDNA quantification.
cccDNA was quantified as previously described (55). The cccDNA quantity was normalized for the amount of nuclear extracted DNA.
Pre-S1 peptide time of addition.
HBV-infected PHHs were treated with 0.5 μM pre-S1 peptide. Cells were incubated with the pre-S1 peptide either during the inoculation for 24 h or from day 1, day 2, or day 5 onward until the end of the infection experiment (day 8). At the end of the infection experiment the percentages of HBsAg- and HBcAg-positive cells were quantified by immunofluorescence and analyzed by high-content imaging (Opera Phenix; PerkinElmer) (magnification, ×20). The percentage of infected cells was quantified by automated image analysis (Phaedra). Percent inhibition was calculated relative to the percentages of HBsAg- and HBcAg-positive cells in the untreated controls.
Quantification of NVR 3-778 by LC-MS/MS.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was carried out on an API4000 Sciex triple quad instrument (ABSciex, Toronto, CA), which was coupled to a high-performance liquid chromatography (HPLC) system (Shimadzu). The chromatographic separation was carried out using a reverse-phase HPLC column (XBridge C18 column; 3.5-μm pore size; 50 by 2.1 mm). Mobile phases consisted of 0.1% formic acid (solvent A) and methanol (solvent B), and a gradient elution at 50°C was performed (70% solvent A and 30% solvent B starting conditions, to 98% solvent B in 1.5 min, isocratic hold at 98% solvent B for 1.2 min, and reequilibration to 30% solvent B in 1.3 min). Total run time was 4.0 min, and a flow rate of 0.6 ml/min was used. The MS was operated in the negative ion mode using a TurboIonSpray interface (electrospray ionization) and was optimized for the quantification of NVR 3-778 (multiple reaction monitoring [MRM] transition 431.1 → 172). NVR 3-778 concentrations in unknown samples were calculated by interpolation from the standard curve.
Plasma protein binding.
The free fraction of NVR 3-778 was determined by equilibrium dialysis. To this end, a Dianorm system with identical macro-1 Teflon cells and Spectra/PorRC 2 dialysis membranes (molecular weight [MW] cutoff of 12 to 14 kDa) was used. NVR 3-778 diluted in the different HCMs or in human plasma was subjected to equilibrium dialysis against a Sörenson phosphate buffer (0.067 M, pH 7.17) at 37°C for 4 h. After dialysis, the contents of the two compartments of the dialysis cells were collected separately, and the concentrations of NVR 3-778 in the different HCM, human plasma, and corresponding buffer samples were determined by LC-MS/MS. The free fraction of NVR 3-778 (fu) was calculated as the ratio of the unbound concentration (Cu) to the total concentration in medium after dialysis (CED). The percentage of unbound compound was calculated as fu × 100, where fu = Cu/CED.
Determination of the in vitro pharmacokinetics of NVR 3-778.
Mock-infected PHHs were incubated for 1, 24, 48, and 72 h with 1 μM NVR 3-778, and the parent drug was quantified by LC/MS-MS in both hepatocyte lysates and supernatant samples. The cellular concentration of the parent drug was normalized for the number of plated PHHs and for the hepatocyte volume (2.28 μl/106 cells) (56). The total cell-associated drug accumulation (Kp) ratio was calculated as Kp = Ccell/Cmedium, where Cmedium is the concentration (in micromolars) of the parent drug in the medium, and Ccell is the cellular parent drug concentration (in micromolars).
The predicted liver-to-medium ratio represents the predicted amount (A) of NVR 3-778 per gram of liver tissue (nanograms/gram of liver tissue) over the amount of NVR 3-778 in the medium (nanograms/milliliter), taking the hepatocellularity of 117.5 million hepatocytes per gram of liver tissue into account (57): liver/medium = Acell (117.5)/Amedium, where Acell is the amount of NVR 3-778 per million cells (nanograms/million) and Amedium is the concentration of NVR 3-778 per milliliter of culture medium (nanograms/milliliter).
The elimination rate constant (k) was calculated from the slope of the linear regression from the natural log concentration remaining in the incubation medium, ln(Cmedium), versus the incubation time (t): ln(Cmedium) = −kt.
The intrinsic apparent hepatic clearance (CLint,app) was calculated according to the following equation: CLint,app = kv/N, where k is the elimination rate constant (per minute), V is incubation volume (150 μl), and N is the number of hepatocytes (0.080 × 106 cells) in the incubation.
The intrinsic clearance was then further corrected for the free fraction of NVR 3-778 in the incubation medium (fu) determined by equilibrium dialysis: CLint = CLint,app/fu.
The average exposure (Cavg) of PHHs to NVR 3-778 was determined by: Cavg = AUC/t, where AUC is the area under the concentration-time curve and t is the incubation time (72 h).
Statistics.
Data values represent means ± standard errors of the means (SEM) and were analyzed by a two-tailed Student's t test. P values of <0.05, <0.01, and <0.001, were considered statistically significant, as indicated on the figures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yannick Debing, Jan Martin Berke, Lore Verschueren, Christel van den Eynde, Wendy Mostmans, and other members of the infectious diseases team at Janssen Pharmaceuticals for providing help with the experiments and useful discussions. We thank Angela Lam for reviewing the manuscript. We thank Ivo Goris for help with the plasma protein binding experiments. We thank Ellen Van Damme and Frank Jacobs for setting up the hepatocyte infection model. We thank Tinne Huybrechts, Lieve Dillen, and Chris Pauwels for assistance with the bioanalysis experiments.
All authors are full-time employees of Janssen Pharmaceuticals.
N.H. conceived the study, generated the data, and wrote the manuscript. T.V. contributed to the AlphaLisa experiments. J.V., O.L., M.M., F.P., and J.S. provided critical input in the study design and data analysis and contributed to the writing of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01497-18.
REFERENCES
- 1.Tsai KN, Kuo CF, Ou JJ. 2018. Mechanisms of hepatitis B virus persistence. Trends Microbiol 26:33–42. doi: 10.1016/j.tim.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang TJ. 2009. Hepatitis B: the virus and disease. Hepatology 49:S13–S21. doi: 10.1002/hep.22881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seeger C, Mason WS. 2015. Molecular biology of hepatitis B virus infection. Virology 479–480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zlotnick A, Venkatakrishnan B, Tan Z, Lewellyn E, Turner W, Francis S. 2015. Core protein: a pleiotropic keystone in the HBV lifecycle. Antiviral Res 121:82–93. doi: 10.1016/j.antiviral.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo YH, Li YN, Zhao JR, Zhang J, Yan Z. 2011. HBc binds to the CpG islands of HBV cccDNA and promotes an epigenetic permissive state. Epigenetics 6:720–726. doi: 10.4161/epi.6.6.15815. [DOI] [PubMed] [Google Scholar]
- 6.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Huser N, Durantel D, Liang TJ, Munk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. 2014. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole AG. 2016. Modulators of HBV capsid assembly as an approach to treating hepatitis B virus infection. Curr Opin Pharmacol 30:131–137. doi: 10.1016/j.coph.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Berke JM, Dehertogh P, Vergauwen K, Van Damme E, Mostmans W, Vandyck K, Pauwels F. 2017. Capsid assembly modulators have a dual mechanism of action in primary human hepatocytes infected with hepatitis B virus. Antimicrob Agents Chemother 61:e00560-17. doi: 10.1128/AAC.00560-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stray SJ, Zlotnick A. 2006. BAY 41-4109 has multiple effects on Hepatitis B virus capsid assembly. J Mol Recognit 19:542–548. doi: 10.1002/jmr.801. [DOI] [PubMed] [Google Scholar]
- 10.Pei Y, Wang C, Yan SF, Liu G. 2017. Past, current, and future developments of therapeutic agents for treatment of chronic hepatitis B virus infection. J Med Chem 60:6461–6479. doi: 10.1021/acs.jmedchem.6b01442. [DOI] [PubMed] [Google Scholar]
- 11.Diab A, Foca A, Zoulim F, Durantel D, Andrisani O. 2018. The diverse functions of the hepatitis B core/capsid protein (HBc) in the viral life cycle: implications for the development of HBc-targeting antivirals. Antiviral Res 149:211–220. doi: 10.1016/j.antiviral.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani N, Cole AG, Phelps JR, Ardzinski A, Cobarrubias KD, Cuconati A, Dorsey BD, Evangelista E, Fan K, Guo F, Guo H, Guo JT, Harasym TO, Kadhim S, Kultgen SG, Lee ACH, Li AHL, Long Q, Majeski SA, Mao R, McClintock KD, Reid SP, Rijnbrand R, Snead NM, Micolochick Steuer HM, Stever K, Tang S, Wang X, Zhao Q, Sofia MJ. 2018. Preclinical profile of AB-423, an inhibitor of hepatitis B virus pregenomic RNA encapsidation. Antimicrob Agents Chemother 62:e00082-18. doi: 10.1128/AAC.00082-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlicksup CJ, Wang JC, Francis S, Venkatakrishnan B, Turner WW, VanNieuwenhze M, Zlotnick A. 2018. Hepatitis B virus core protein allosteric modulators can distort and disrupt intact capsids. Elife 7:e31473. doi: 10.7554/eLife.31473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campagna MR, Liu F, Mao R, Mills C, Cai D, Guo F, Zhao X, Ye H, Cuconati A, Guo H, Chang J, Xu X, Block TM, Guo JT. 2013. Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids. J Virol 87:6931–6942. doi: 10.1128/JVI.00582-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feld JJ, Colledge D, Sozzi V, Edwards R, Littlejohn M, Locarnini SA. 2007. The phenylpropenamide derivative AT-130 blocks HBV replication at the level of viral RNA packaging. Antiviral Res 76:168–177. doi: 10.1016/j.antiviral.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 16.Yuen MF, Kim DJ, Weiler F, Chan HLY, Lalezari J, Hwang SG, Nguyen T, Liaw S, Brown N, Klumpp K, Flores L, Hartman G, Gane EJ. 2015. Phase 1b efficacy and safety of NVR 3-778, a first-in-class HBV core inhibitor, in HBeAg-positive patients with chronic HBV infection, abstr LB-10. Abstr 66th Annu Meet Am Assoc Study Liver Dis (AASLD), 13 to 17 November 2015, Boston, MA. [Google Scholar]
- 17.Yuen MF, Kim DJ, Weiler F, Chan HLY, Lalezari J, Hwang SG, Nguyen T, Liaw S, Brown N, Klumpp K, Flores L, Hartman G, Gane EJ. 2016. NVR 3-778, a first-in-class HBV core inhibitor, alone and in combination with pegylated-interferon (peg-IFN alpha-2a), in treatment-naïve HBeAg-positive patients: early reductions in HBV DNA and HBeAg. J Hepatol 64:S210–S211. doi: 10.1016/S0168-8278(16)00175-6. [DOI] [Google Scholar]
- 18.Sells MA, Chen ML, Acs G. 1987. Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A 84:1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu L, Cheng YC. 2000. Characterization of novel human hepatoma cell lines with stable hepatitis B virus secretion for evaluating new compounds against lamivudine- and penciclovir-resistant virus. Antimicrob Agents Chemother 44:3402–3407. doi: 10.1128/AAC.44.12.3402-3407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, King RW. 1997. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother 41:1715–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westerink WM, Schoonen WG. 2007. Cytochrome P450 enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol In Vitro 21:1581–1591. doi: 10.1016/j.tiv.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Wilkening S, Stahl F, Bader A. 2003. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab Dispos 31:1035–1042. doi: 10.1124/dmd.31.8.1035. [DOI] [PubMed] [Google Scholar]
- 23.Schulze A, Mills K, Weiss TS, Urban S. 2012. Hepatocyte polarization is essential for the productive entry of the hepatitis B virus. Hepatology 55:373–383. doi: 10.1002/hep.24707. [DOI] [PubMed] [Google Scholar]
- 24.Galle PR, Hagelstein J, Kommerell B, Volkmann M, Schranz P, Zentgraf H. 1994. In vitro experimental infection of primary human hepatocytes with hepatitis B virus. Gastroenterology 106:664–673. doi: 10.1016/0016-5085(94)90700-5. [DOI] [PubMed] [Google Scholar]
- 25.Shlomai A, Schwartz RE, Ramanan V, Bhatta A, de Jong YP, Bhatia SN, Rice CM. 2014. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc Natl Acad Sci U S A 111:12193–12198. doi: 10.1073/pnas.1412631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soars MG, McGinnity DF, Grime K, Riley RJ. 2007. The pivotal role of hepatocytes in drug discovery. Chem Biol Interact 168:2–15. doi: 10.1016/j.cbi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Hewitt NJ, Lechon MJ, Houston JB, Hallifax D, Brown HS, Maurel P, Kenna JG, Gustavsson L, Lohmann C, Skonberg C, Guillouzo A, Tuschl G, Li AP, LeCluyse E, Groothuis GM, Hengstler JG. 2007. Primary hepatocytes: current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metab Rev 39:159–234. doi: 10.1080/03602530601093489. [DOI] [PubMed] [Google Scholar]
- 28.Bohnert T, Gan LS. 2013. Plasma protein binding: from discovery to development. J Pharm Sci 102:2953–2994. doi: 10.1002/jps.23614. [DOI] [PubMed] [Google Scholar]
- 29.Smith DA, Di L, Kerns EH. 2010. The effect of plasma protein binding on in vivo efficacy: misconceptions in drug discovery. Nat Rev Drug Discov 9:929–939. doi: 10.1038/nrd3287. [DOI] [PubMed] [Google Scholar]
- 30.Adkins JN, Varnum SM, Auberry KJ, Moore RJ, Angell NH, Smith RD, Springer DL, Pounds JG. 2002. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol Cell Proteomics 1:947–955. doi: 10.1074/mcp.M200066-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Blain PG, Mucklow JC, Rawlins MD, Roberts DF, Routledge PA, Shand DG. 1985. Determinants of plasma alpha 1-acid glycoprotein (AAG) concentrations in health. Br J Clin Pharmacol 20:500–502. doi: 10.1111/j.1365-2125.1985.tb05107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mora-Peris B, Watson V, Vera JH, Weston R, Waldman AD, Kaye S, Khoo S, Mackie NE, Back D, Winston A. 2014. Rilpivirine exposure in plasma and sanctuary site compartments after switching from nevirapine-containing combined antiretroviral therapy. J Antimicrob Chemother 69:1642–1647. doi: 10.1093/jac/dku018. [DOI] [PubMed] [Google Scholar]
- 33.Sims KD, Lemm J, Eley T, Liu M, Berglind A, Sherman D, Lawitz E, Vutikullird AB, Tebas P, Gao M, Pasquinelli C, Grasela DM. 2014. Randomized, placebo-controlled, single-ascending-dose study of BMS-791325, a hepatitis C virus (HCV) NS5B polymerase inhibitor, in HCV genotype 1 infection. Antimicrob Agents Chemother 58:3496–3503. doi: 10.1128/AAC.02579-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taverna M, Marie AL, Mira JP, Guidet B. 2013. Specific antioxidant properties of human serum albumin. Ann Intensive Care 3:4. doi: 10.1186/2110-5820-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heslop JA, Rowe C, Walsh J, Sison-Young R, Jenkins R, Kamalian L, Kia R, Hay D, Jones RP, Malik HZ, Fenwick S, Chadwick AE, Mills J, Kitteringham NR, Goldring CE, Kevin Park B. 2017. Mechanistic evaluation of primary human hepatocyte culture using global proteomic analysis reveals a selective dedifferentiation profile. Arch Toxicol 91:439–452. doi: 10.1007/s00204-016-1694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schieck A, Schulze A, Gahler C, Muller T, Haberkorn U, Alexandrov A, Urban S, Mier W. 2013. Hepatitis B virus hepatotropism is mediated by specific receptor recognition in the liver and not restricted to susceptible hosts. Hepatology 58:43–53. doi: 10.1002/hep.26211. [DOI] [PubMed] [Google Scholar]
- 37.Chiba M, Ishii Y, Sugiyama Y. 2009. Prediction of hepatic clearance in human from in vitro data for successful drug development. AAPS J 11:262–276. doi: 10.1208/s12248-009-9103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyauchi S, Masuda M, Kim SJ, Tanaka Y, Lee KR, Iwakado S, Nemoto M, Sasaki S, Shimono K, Tanaka Y, Sugiyama Y. 2018. The phenomenon of albumin-mediated hepatic uptake of organic anion transport polypeptide substrates: prediction of the in vivo uptake clearance from the in vitro uptake by isolated hepatocytes using a facilitated-dissociation model. Drug Metab Dispos 46:259–267. doi: 10.1124/dmd.117.077115. [DOI] [PubMed] [Google Scholar]
- 39.Di L, Feng B, Goosen TC, Lai Y, Steyn SJ, Varma MV, Obach RS. 2013. A perspective on the prediction of drug pharmacokinetics and disposition in drug research and development. Drug Metab Dispos 41:1975–1993. doi: 10.1124/dmd.113.054031. [DOI] [PubMed] [Google Scholar]
- 40.Baker M, Parton T. 2007. Kinetic determinants of hepatic clearance: plasma protein binding and hepatic uptake. Xenobiotica 37:1110–1134. doi: 10.1080/00498250701658296. [DOI] [PubMed] [Google Scholar]
- 41.Liu X, Wright M, Hop CE. 2014. Rational use of plasma protein and tissue binding data in drug design. J Med Chem 57:8238–8248. doi: 10.1021/jm5007935. [DOI] [PubMed] [Google Scholar]
- 42.Lai MT, Munshi V, Touch S, Tynebor RM, Tucker TJ, McKenna PM, Williams TM, DiStefano DJ, Hazuda DJ, Miller MD. 2009. Antiviral activity of MK-4965, a novel nonnucleoside reverse transcriptase inhibitor. Antimicrob Agents Chemother 53:2424–2431. doi: 10.1128/AAC.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrail-Tran A, Mentre F, Cosson C, Piketty C, Chazallon C, Gerard L, Girard PM, Taburet AM. 2010. Influence of alpha-1 glycoprotein acid concentrations and variants on atazanavir pharmacokinetics in HIV-infected patients included in the ANRS 107 trial. Antimicrob Agents Chemother 54:614–619. doi: 10.1128/AAC.00797-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Owens CM, Brasher BB, Polemeropoulos A, Rhodin MH, McAllister N, Peng X, Wang C, Ying L, Cao H, Lawitz E, Poordad F, Rondon J, Box TD, Zeuzem S, Buggisch P, Lin K, Qiu YL, Jiang L, Colvin R, Or YS. 2016. Preclinical profile and clinical efficacy of a novel hepatitis C virus NS5A inhibitor, EDP-239. Antimicrob Agents Chemother 60:6207–6215. doi: 10.1128/AAC.00808-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin TI, Lenz O, Fanning G, Verbinnen T, Delouvroy F, Scholliers A, Vermeiren K, Rosenquist A, Edlund M, Samuelsson B, Vrang L, de Kock H, Wigerinck P, Raboisson P, Simmen K. 2009. In vitro activity and preclinical profile of TMC435350, a potent hepatitis C virus protease inhibitor. Antimicrob Agents Chemother 53:1377–1385. doi: 10.1128/AAC.01058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molla A, Vasavanonda S, Kumar G, Sham HL, Johnson M, Grabowski B, Denissen JF, Kohlbrenner W, Plattner JJ, Leonard JM, Norbeck DW, Kempf DJ. 1998. Human serum attenuates the activity of protease inhibitors toward wild-type and mutant human immunodeficiency virus. Virology 250:255–262. doi: 10.1006/viro.1998.9383. [DOI] [PubMed] [Google Scholar]
- 47.Brunner D, Frank J, Appl H, Schoffl H, Pfaller W, Gstraunthaler G. 2010. Serum-free cell culture: the serum-free media interactive online database. ALTEX 27:53–62. doi: 10.14573/altex.2010.1.53. [DOI] [PubMed] [Google Scholar]
- 48.Thung SN, Wang DF, Fasy TM, Hood A, Gerber MA. 1989. Hepatitis B surface antigen binds to human serum albumin cross-linked by transglutaminase. Hepatology 9:726–730. doi: 10.1002/hep.1840090512. [DOI] [PubMed] [Google Scholar]
- 49.Pontisso P, Alberti A, Schiavon E, Tremolada F, Bortolotti F, Realdi G. 1983. Receptors for polymerized human serum albumin on hepatitis B virus particles detected by radioimmunoassay: changes in receptor activity in serum during acute and chronic infection. J Virol Methods 6:151–159. doi: 10.1016/0166-0934(83)90027-7. [DOI] [PubMed] [Google Scholar]
- 50.Michel ML, Pontisso P, Sobczak E, Malpiece Y, Streeck RE, Tiollais P. 1984. Synthesis in animal cells of hepatitis B surface antigen particles carrying a receptor for polymerized human serum albumin. Proc Natl Acad Sci U S A 81:7708–7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Summers J, Smith PM, Horwich AL. 1990. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J Virol 64:2819–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lentz TB, Loeb DD. 2011. Roles of the envelope proteins in the amplification of covalently closed circular DNA and completion of synthesis of the plus-strand DNA in hepatitis B virus. J Virol 85:11916–11927. doi: 10.1128/JVI.05373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lam A, Espiritu C, Flores L, Hartman G, Klumpp K. 2015. Effect of the combination of the HBV core inhibitor NVR 3-778 with nucleoside analogs or other HBV core inhibitors on the inhibition of HBV DNA replication in HepG2.2.15 cells. J Hepatol 62(Suppl 2):S559. doi: 10.1016/S0168-8278(15)30845-X. [DOI] [Google Scholar]
- 54.Klumpp K, Shimada T, Allweiss L, Volz T, Lutgehetmann M, Hartman G, Flores OA, Lam AM, Dandri M. 2018. Efficacy of NVR 3-778, Alone and In Combination With Pegylated Interferon, versus Entecavir In uPA/SCID Mice With Humanized Livers and HBV Infection. Gastroenterology 154:652–662.e8. doi: 10.1053/j.gastro.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 55.Lam AM, Ren S, Espiritu C, Kelly M, Lau V, Zheng L, Hartman GD, Flores OA, Klumpp K. 2017. Hepatitis B virus capsid assembly modulators, but not nucleoside analogs, inhibit the production of extracellular pregenomic RNA and spliced RNA variants. Antimicrob Agents Chemother 61:e00680-17. doi: 10.1128/AAC.00680-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshikado T, Toshimoto K, Nakada T, Ikejiri K, Kusuhara H, Maeda K, Sugiyama Y. 2017. Comparison of methods for estimating unbound intracellular-to-medium concentration ratios in rat and human hepatocytes using statins. Drug Metab Dispos 45:779–789. doi: 10.1124/dmd.116.074823. [DOI] [PubMed] [Google Scholar]
- 57.Ellens H, Johnson M, Lawrence SK, Watson C, Chen L, Richards-Peterson LE. 2017. Prediction of the transporter-mediated drug-drug interaction potential of dabrafenib and its major circulating metabolites. Drug Metab Dispos 45:646–656. doi: 10.1124/dmd.116.073932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.