Fig. 2. DNA substrate inhibition of UvrD-catalyzed DNA unwinding is relieved by MutL.
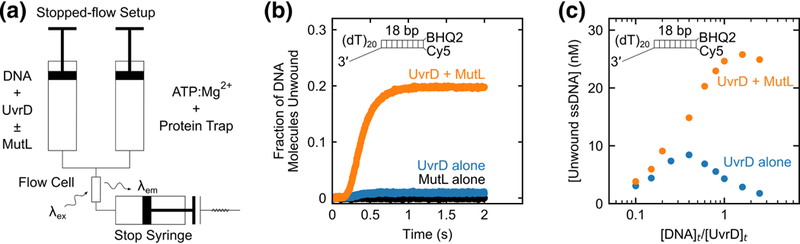
a, Schematic of the stopped-flow DNA unwinding experiment. In one syringe UvrD is pre-incubated with DNA labeled with Cy5 and BHQ2, with or without MutL in buffer T at 25 °C. UvrD-DNA-(±MutL) complex is rapidly mixed with ATP, MgCl2, and a protein trap to initiate DNA unwinding. DNA strand separation is accompanied by an enhancement in Cy5 fluorescence signal. b, DNA unwinding time courses from experiments performed at 125 nM 3’-(dT)20-ds18-BHQ2/Cy5 with 50 nM UvrD alone (blue), 625 nM MutL dimer alone (black) or 50 nM UvrD plus 625 nM MutL dimer (orange). c, Multiple DNA unwinding experiments were performed at constant 50 nM UvrD over a range of DNA concentrations in the presence (orange circles) or in the absence (blue circles) of saturating MutL (5-fold molar excess of MutL dimer over DNA) and the total unwinding amplitudes were plotted as a function of [DNA]t/[UvrD]t.
