Fig. 5. A single MutL dimer can activate a UvrD monomer.
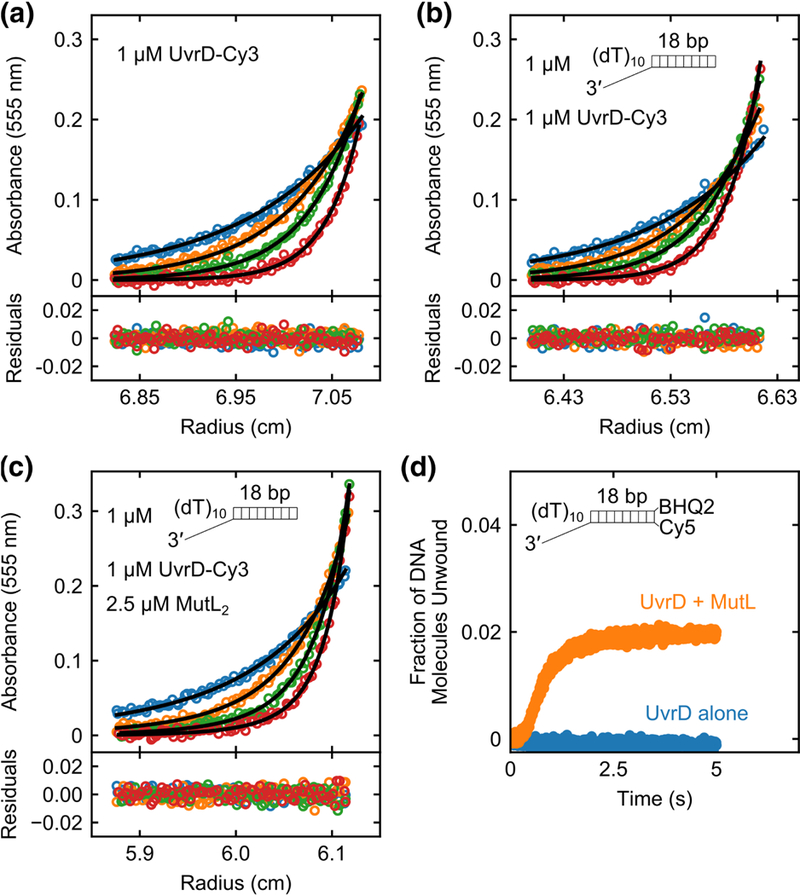
Sedimentation equilibrium experiments monitoring the Cy3 absorbance of UvrD-Cy3 at 555 nm were performed in buffer M20/20 at 25 °C. (a) UvrD-Cy3 alone (1 μM) shows a single species corresponding to a monomer at four rotor speeds (12k rpm (blue); 15k rpm (orange); 18k rpm (green); 22k rpm (red)). (b) UvrD-Cy3 (1 μM) plus 3’-(dT)10-ds18 (1 μM) shows a single species corresponding to a UvrD-Cy3 monomer-DNA complex at four rotor speeds (12k rpm (blue); 15k rpm (orange); 18k rpm (green); 22k rpm (red)). (c) UvrD- Cy3 (1 μM), 3’-(dT)10-ds18 (1 μM) and MutL (2.5 μM dimer) mixture shows two species containing UvrD-Cy3 at four rotor speeds (9k rpm (blue); 12k rpm (orange); 15k rpm (green); 18k rpm (red)). The two species correspond to a (UvrD-Cy3 monomer-DNA) complex and a (MutL dimer-UvrD-Cy3 monomer-DNA) complex. Smooth curves are simulations using best fit parameters with residuals shown below the plots as described in the text. d, Stopped-flow DNA unwinding time courses with 100 nM 3’-(dT)10-ds18- BHQ2/Cy5 DNA in buffer M20/20 at 25 °C were performed with 100 nM UvrD alone (blue) or 100 nM UvrD plus 250 nM MutL dimer (orange).
