Figure 1.
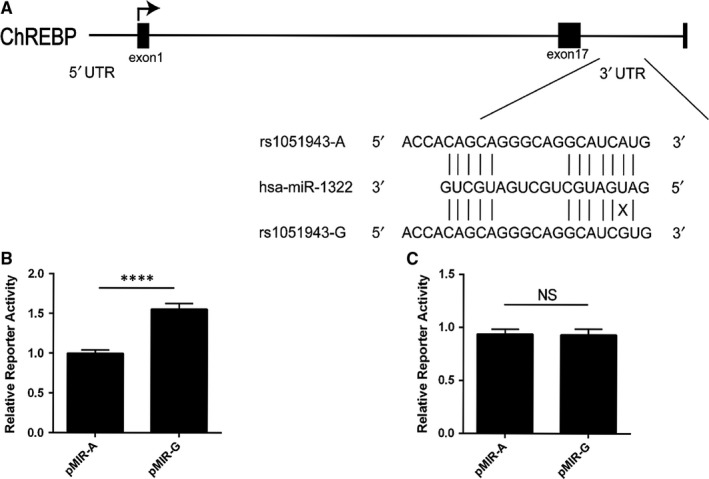
Functional analysis of rs1051943 A/G polymorphism. A, ChREBP gene structure and the rs1051943 polymorphism located in the 3’‐UTR at the miR‐1322 binding site. The rs1051943 polymorphism is a A–G change (mRNA sequence as reference) located in the predicted binding site of miR‐1322. According to Watson‐Crick mode, rs1051943 A allele base‐paired with U allele at the miRNA seed sequence (shown with a vertical line), whereas G allele did not (shown with a cross). B, C, Effects of rs1051943A/G allele on ChREBP 3’‐UTR constructs activity in HepG2 cells (B) and 293T cells (C). ChREBP 3’‐UTR reporter activity is expressed relative to the co‐transfected Renilla luciferase activity. Three independent experiments containing six replicates were performed in each cell line. For each comparison, mutant construct was compared with the wild‐type construct. All values are mean ± SEM of three independent experiments with six replicates. P < 0.0001 (****), NS (no significance)
