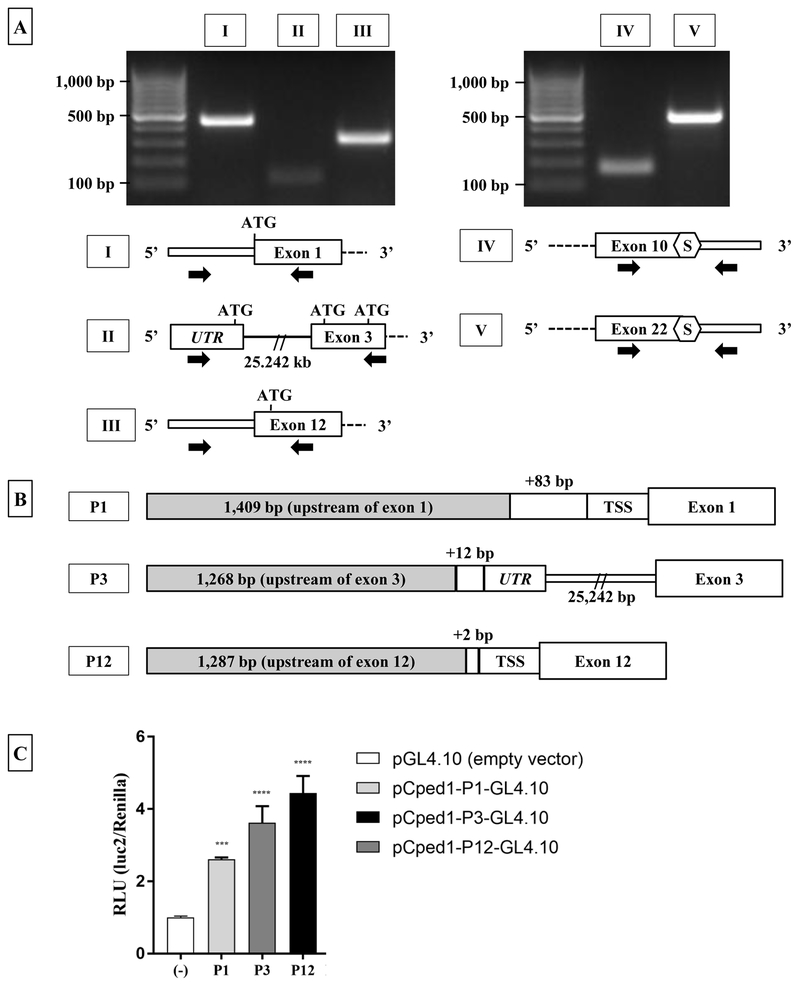
Figure 2: Calvaria express multiple Cped1 transcripts with alternate 5’- and 3’-UTR, and utilize alternate promoters for transcription.
A) RT-PCR of Cped1 transcripts demonstrates the expression of alternate 5’- and 3’-UTRs. Thick and thin rectangles represent exon and UTRs, respectively, and solid lines represent introns. White hexagons labeled ‘S’ represent predicted stop codons as identified by analysis of nucleotide sequence. Black arrows represent the approximate target location of forward and reverse primers for PCR of Cped1 transcripts. B) Predicted promoter regions upstream of exon 1 (P1), exon 3 (P3), and exon 12 (P12) are transcriptionally active in MC3T3-E1 pre-osteoblasts during osteogenic differentiation. Fold change between luciferase constructs P1, P3, and P12 vs. empty vector (pGL4.10) was detected. Data is represented as change in relative light units (RLU) of firefly luciferase normalized to renilla luciferase 24 hours post-transfection. Results are expressed as mean +/− SD, N=3. TSS: translational start site. Statistical differences were detected via one-way ANOVA and Dunnett’s multiple comparisons test. ***P < 0.001, ****P < 0.0001