Figure 3.
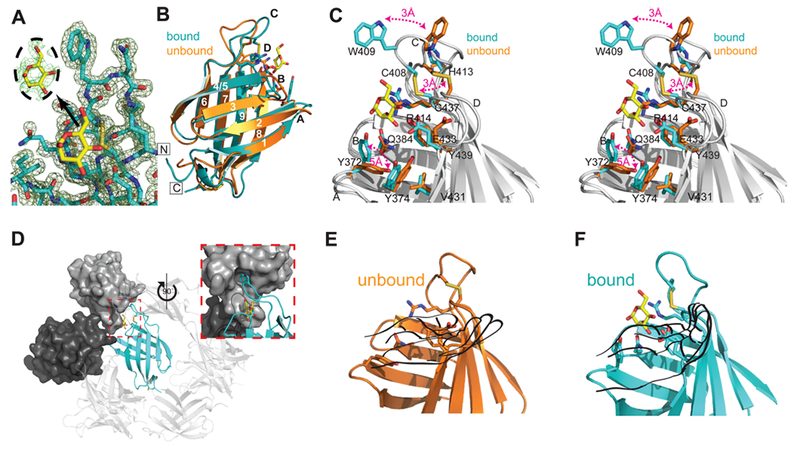
Crystal structure of the S. pombe GIIβ MRH domain bound to mannose. (A) Final 2Fo – Fc map contoured at 1σ with the Fo – Fc map for the ligand contoured at 2σ displayed in the inset. (B) Crystal structure of the MRH domain bound (PDB entry 4XQM) (cyan) to mannose (yellow) in comparison to the NMR structure of the unbound (orange) protein (PDB entry 2LVX). Loops A–D and β-strands 1–9 are labeled. The N- and C-termini are labeled and boxed. (C) Close-up stereoview of the binding pocket of the superimposed bound (cyan) and unbound (orange) structures with residues shown as sticks. Three major changes between the apo and bound structures are highlighted with red double arrows. Additionally, when ligand binds, the binding pocket narrows as the hydroxyl group of Y439 moves toward the N-terminal β-sheet by slightly more than 1 Å, and Tyr374 rotates ~45° so that the plane of its phenolic ring is oriented perpendicular to the side chain of V431. (D) Crystallographic packing of GIIβ MRH (cyan ribbon). The molecular surface of the cystallographic neighbor that contacts both W409 and the ligand is colored dark gray. The molecule that further occludes the binding region is colored light gray. The inset is rotated 90° with reference to a main panel. Ribbon diagram of GIIβ MRH domain in the absence (E) and presence (F) of mannose (yellow). Although in the unbound NMR structure previously reported33 the chemical shifts for these nitrogen atoms and their associated protons could not be determined, the energy-minimized structure consistently used the fully extended rotamer of R414. Figures were generated using PyMOL.51
