Figure 7.
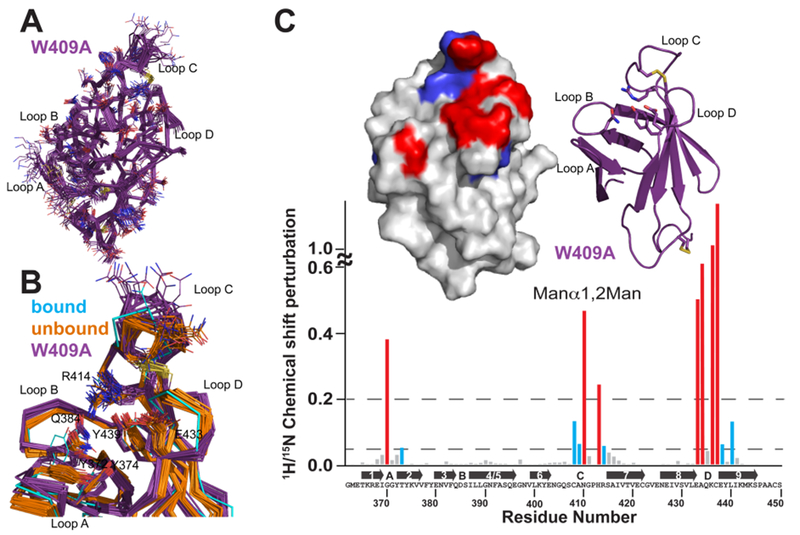
Solution structure of the S. pombe GIIβ MRH domain mutant W409A. (A) Superimposition of the polypeptide backbone heavy atoms of the 20 lowest-energy conformers of the ensemble representing the NMR structure of W409A. The side chains of the ensemble members are shown as lines. The chemical shifts for the first five N-terminal residues were not determined and for the sake of clarity are not shown. (B) Superimposition of the crystallographically bound model with the ensemble members of the S. pombe GIIβ MRH domain in the ligand-unbound state for the wild type (orange) and the W409A mutant (purple). Residues discussed in the text are represented by lines. A close-up view of the binding pocket is shown. (C) Spectrum of the GIIβ-MRH W409A mutant (0.1 mM) collected in the presence and absence of 50 mM Manβ1,2Man. 15N–1H chemical shift perturbations are plotted vs residue number for the protein. Secondary structural elements are listed above the sequence. Missing values correspond to proline residues or residues not observed in the 15N–1H HSQC spectra. The top left inset depicts chemical shift mapping on the structure surface. The surface is colored red for amino acids with the largest chemical shift perturbations (>0.2) and blue for those with more moderate (0.05) perturbations. The top right inset is a ribbon diagram for the molecule in the same orientation as the surface representation. The four essential amino acids are shown as sticks.
