Abstract
Serine hydrolases play diverse roles in regulating host-pathogen interactions in a number of organisms, yet few have been characterized in the human pathogen Staphylococcus aureus. Here, we describe a chemical proteomic screen that identified 10 previously uncharacterized S. aureus serine hydrolases that mostly lack human homologues. We termed these enzymes Fluorophosphonate-binding hydrolases (FphA-J). One hydrolase, FphB, can process short fatty acid esters, exhibits increased activity in response to host cell factors, is located predominantly on the bacterial cell surface in a subset of cells, and is concentrated in the division septum. Genetic disruption of the fphB gene confirms that the enzyme is dispensable for bacterial growth in culture but crucial for establishing infection in distinct sites in vivo. A selective small molecule inhibitor of FphB effectively reduces infectivity in vivo, suggesting that it may be a viable therapeutic target for the treatment or management of Staphylococcus infections.
Introduction
The bacterium Staphylococcus aureus is a highly significant human pathogen and a major cause of bacterial infections worldwide1. While this bacterium is found as part of the commensal skin and mucosal microbiome in about 30% of the human population, it is usually held in check by the physical barrier of the skin. However, upon breach of host defenses, S. aureus can disseminate systemically, leading to life-threatening conditions such as endocarditis, osteomyelitis, meningitis and sepsis2. S. aureus often forms highly robust biofilms in which the bacterium populates synthetic surfaces, e.g. on prosthetic devices, surrounding itself in a biomolecular matrix that is largely impermeable to the immune system and many antibiotics, causing infections that are complicated and costly to monitor and treat3. Finally, an increased prevalence of community-acquired infections with antibiotic-resistant strains2 is a further reason why S. aureus remains a major human health threat that requires new methods for rapid detection, treatment and therapy response monitoring.
Hydrolytic enzymes have vital roles for maintaining bacterial homeostasis and survival at the host-pathogen interface4–10 and thus represent potential anti-virulence and anti-infectivity targets11,12. Serine hydrolases are one of the largest and most diverse enzyme classes in eukaryotic and prokaryotic proteomes13. One of the largest subgroups of serine hydrolases, the α,β-hydrolases, are classified as enzymes that play important roles in processing of metabolites, peptides and lipids as a means of controlling cell signaling and metabolism. They also have been proven to be effective drug targets for a variety of diseases14–16, yet the functionalities of α,β-hydrolases in S. aureus remain largely unknown.
Serine hydrolases proceed through an acyl enzyme intermediate during processing of a substrate, thus a range of serine reactive electrophiles can be used to covalently target these enzymes17. In fact, flurophosphonate-based probes have been used to globally profile serine hydrolases in animal tissues11,18, human cell lines19, the pathogenic bacteria Mycobacterium tuberculosis20,21 and Vibrio cholerae 22 and in archaea23. Here, we describe a functional proteomic screen in live S. aureus under biofilm-promoting conditions using the serine-reactive activity-based probe (ABP) fluorophosphonate-tetramethylrhodamine (FP-TMR)12. This screen identified a set of 10 α,β-hydrolase containing enzymes that are expressed in live S. aureus that have little or no homology to host-derived serine hydrolases. Through the identification and use of a covalent inhibitor, development of a highly-selective fluorescent ABP and the use of genetic knock-out strains, we were able to characterize one of these hydrolases as a virulence factor. This enzyme is localized at the bacterial surface, is heterogeneously distributed in the bacterial population, can process short-chain lipid esters, is regulated in response to host-cell derived factors and is important for infection of distinct tissue sites in vivo. Our results demonstrate the power of a focused functional proteomic screen in a relevant human pathogen such as S. aureus to identify a group of hydrolytic enzymes whose functions are likely to be important for various aspects of cellular physiology and host-pathogen interactions. Given their accessibility to modification by chemical probes and lack of human homologues, these enzymes may be promising targets for diagnosis, infection monitoring or treatment of S. aureus infections.
Results
Serine hydrolase activity profiling and inhibitor screen
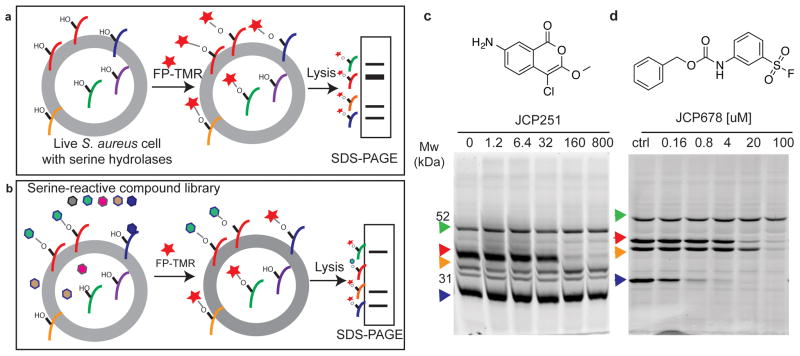
Global profiles of serine hydrolase activity can be generated by treating intact cells with the fluorescent ABP FP-TMR followed by analysis of labeled protein by SDS-PAGE analysis (Fig. 1a). ABPs also allow screening for inhibitors of newly identified enzyme targets without the need to express the enzymes and identify substrates by competition labeling (Fig. 1b). Therefore, we set out to identify novel serine hydrolase targets in S. aureus using the FP-TMR probe. Since the biofilm form of S. aureus is highly clinically relevant, we performed initial activity-based protein profiling (ABPP) with FP-TMR in S. aureus cells (strain ATCC3556) that were grown under biofilm-promoting conditions, removed from the matrix and suspended in broth. This identified multiple prominently labeled enzymes that were resolved by SDS-PAGE analysis of the total cellular lysates (Fig. 1c). Without knowing the identity of these targets, we screened a library of ~500 compounds that were designed to covalently target serine proteases and hydrolases24,25 using the competition labeling method (Supplementary Fig. 1a, Supplementary Table 1). Deconvolution of the original screening mixtures and secondary screening of individual compounds identified potent and selective inhibitors of two of the most efficiently labeled hydrolases (Supplementary Fig. 1b, Supplementary Table 1). Specifically, compound 1, the chloroisocoumarin JCP251, was selective for a ~36 kDa hydrolase target at nanomolar concentrations (Fig. 1c) and compound 2, the sulfonyl fluoride JCP678, showed selectivity for a ~28 kDa hydrolase at low micromolar concentrations (Fig. 1d).
Fig. 1. Identification of serine hydrolases and inhibitors in live S. aureus by competitive ABPP.
a) Schematic of serine hydrolase labeling in live S. aureus with FP-TMR (red star) followed by SDS-PAGE analysis to detected labeled targets. b) Schematic overview of competitive activity-based profiling platform to identify selective inhibitors of individual hydrolases from a library of small molecules (hexagons). c) Chemical structure of JCP251 and SDS-PAGE analysis of S. aureus lysates after live cells were incubated with JCP251 prior to FP-TMR labeling. Arrowheads indicate consistently observed serine hydrolase activities. d) Chemical structure of JCP678 and its FP-TMR competition labeling profile in S. aureus. Experiments shown in c,d were repeated twice with similar results.
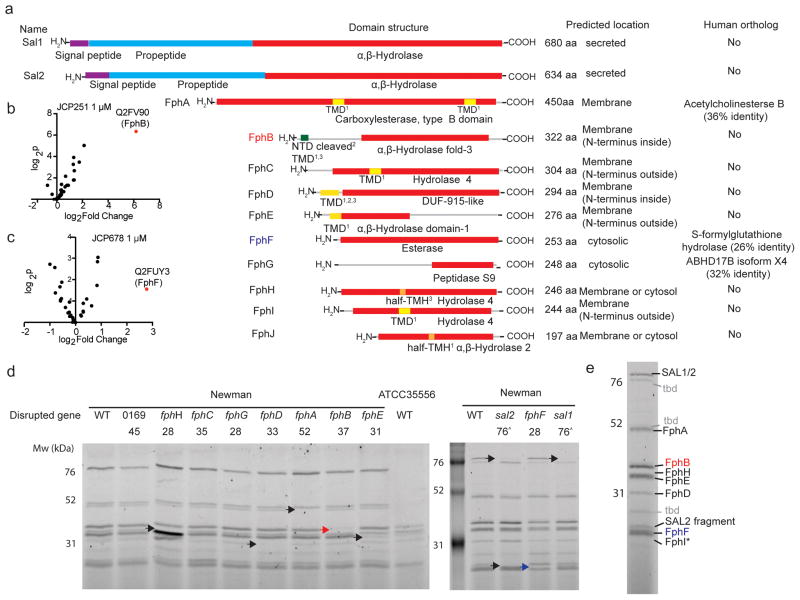
To identify the targets of these inhibitors along with the other probe-labeled S. aureus serine hydrolases, we pretreated live bacteria with JCP251, JCP678 or vehicle control, and then labeled them with the biotin-tagged probe FP-biotin. Cells were lysed, enriched for biotinylated proteins using a streptavidin-resin and samples analyzed by LC-MS/MS (Fig. 2a). Using this strategy, we identified peptides from 12 proteins predicted to contain an α,β-hydrolase domain (Supplementary Table 2, Extended Data Table 1, data are available via ProteomeXchange with identifier PXD009210). All of these proteins, with the exceptions of S. aureus lipases SAL1 and SAL226–28, are poorly annotated with no assigned cellular functions. We then examined the predicted domain structures, cellular location, size and closest human homologs of each of the 12 hydrolases (Fig. 2a). Given that none of the enzymes (except Sal1 and Sal2) have been given names or have identifiable human homologs, we named these 10 enzymes ‘Flurophosphonate-binding serine hydrolases’ (Fphs) following the nomenclature used for penicillin-binding proteins. We assigned sequential letters (A–J) to each enzyme based on the predicted size of the proteins with the largest protein being assigned the letter A. Two of these uncharacterized hydrolases, FphE and FphG, were previously reported (but not named or further studied) in a chemoproteomic study using β-lactam probes that primarily target penicillin-binding proteins6,29. None of the other Fph targets were labeled by the β-lactam probes6. By identifying specific hydrolases whose recovery was blocked by pretreatment with the selective inhibitors, we identified the 36 kDa hydrolase as FphB and the primary target of JCP251 and the 28 kDa protein as FphF and the target of JCP678 (Fig. 2b, c, Supplementary Table 2, Extended Data Table 1, Supplementary Fig. 3).
Fig. 2. LC-MS/MS-based identification of serine hydrolases in S. aureus.
a) Domain structure prediction and bioinformatics analyses of identified hydrolases. Red: α,β-Hydrolase domain (family specification according to Pfam). Purple: signal peptides. Cyan: propeptides. Yellow: transmembrane domain (TMD). Orange: Possible half-transmembrane-helix (half-TMH). Green: Ambiguous N-terminal domain (NTD). The programs predicting the presence of a TMD/TMH are indicated as follows: 1 TmPred, 2Phobius, 3MemBrain. For identification of human orthologs by blastp, the non-redundant protein sequences database for Homo sapiens was queried using the indicated full-length protein sequences. b) Volcano plot of FP-biotin targets in S. aureus ATCC35556 showing change in recovery upon JCP251 and c) JCP678 pretreatment relative to a control. The top hit (highest p value and most significant change in recovery) is labeled. P-values were calculated using a two-tailed t-test on n=3 biologically independent samples for each of the two groups being compared. d) FP-TMR labeling profiles of S. aureus Newman transposon mutant strains with insertions in serine hydrolase genes. Arrowheads indicate labeled proteins disappearing in individual mutant strains (Red arrowhead: FphB, blue arrowhead: FphH). The experiment was performed twice with similar results. e) The primary labeled targets of FP-TMR in S. aureus Newman. Identities of species confirmed by mutational analysis are indicated, * FphI identity is predicted based on molecular weight. ‘tbd’ identity remains unconfirmed
To further confirm the identity of all of the major FP-TMR-labeled hydrolases, we obtained strains from the Nebraska Transposon Mutant Library30 with insertions in individual fph genes. We transduced these mutants into the well-characterized S. aureus Newman lab strain background and performed FP-TMR labeling experiments to correlate individual hydrolases with probe-labeled proteins by SDS-PAGE analysis (Fig. 2d). For transposon mutants strains deficient in either Sal1/2, FphA, FphB, FphE, FphF or FphH, we observed loss of a labeled species of the corresponding molecular weight from the FP-TMR labeled proteome. For the fphD mutant we observed loss of labeling of a hydrolase of <30 kDa, which is smaller than the expected size of 33 kDa, but is consistent with a previous observation6. Among the available transposon mutant strains, we were not able to identify FphC and FphG as probe-labeled bands in the SDS-PAGE gel. This might be due to the low abundance of these enzymes in combination with lower sensitivity and limited resolution of the SDS-PAGE- compared to the MS-based assay, as well as different activity and permeability of the biotinylated and fluorescent probes. Interestingly, the FphH-transposon mutant displayed elevated labeling of several other serine hydrolase activities (e.g. FphE and FphD) suggesting a possible level of functional redundancy and/or compensation among these enzymes (Fig. 2d). Ultimately, we were able to identify 8 Fphs and Sal1/2 in the SDS-PAGE experiment, as well as three additional labeled proteins whose identities have yet to be confirmed (Fig. 2e).
Biochemical analysis of S. aureus FphB
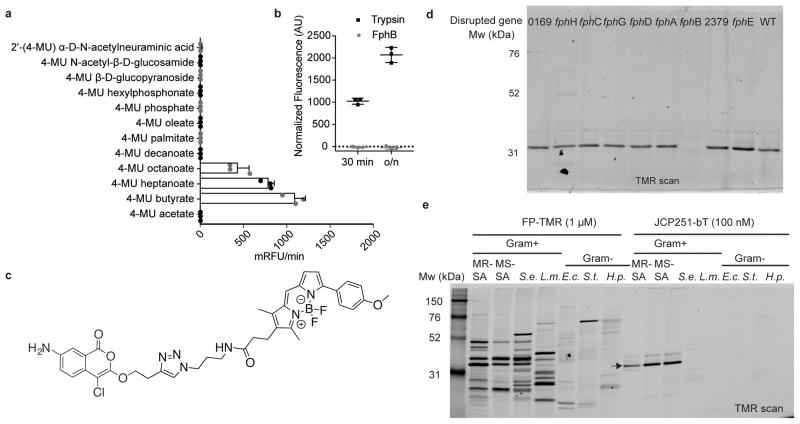
Although we identified 10 serine hydrolases in our proteomic screen that each likely serve important functions to the bacteria, we focused our initial efforts on FphB due to its overall high activity in cells, lack of prior characterization and the overall potency and selectivity of the JCP251 lead inhibitor. FphB is predicted to be a 322 amino acid protein of 36.8 kDa size (Fig. 2a). Bioinformatic analyses suggest FphB does not have a classical signal peptide, is not secreted, but may possess a short N-terminal transmembrane domain or cleavage site (Supplementary Table 2). Data mining of previously published transcriptomic datasets revealed that fphB gene transcription was moderately upregulated (1.8 – 2.9-fold) upon exposure to various cell wall-acting antibiotics and antimicrobial peptides (locus tags SA232331,32,33 and SA254934, resp.). Similar upregulations in fphB transcription were found in response to neutrophil-associated azurophilic granules, hydrogen peroxide, hypochlorus acid, (locus tag MW245635 acid shock (locus tagSA2549)36 as well as antibacterial skin fatty acids, (temporary upregulation, locus tag SA2323)37 suggesting a potential role for FphB under stress conditions at the host-pathogen interface. We recombinantly expressed FphB and purified it as the full length, His6- and T7-tagged fusion protein rFphB from E. coli (Supplementary Fig. 4). To determine if the protein was enzymatically active, we tested a panel of commercially available fluorogenic substrates (Fig. 3a). We found that the rFphB protein cleaved fluorogenic esterase substrates, but did not process phosphate, phosphonate, glycosidic substrates or the protease substrate FITC-casein (Fig. 3b). Among the ester substrates tested, it preferred C4 > C7 > C8 fatty acid esters but was unable to cleave C2, C10 or longer fatty acid esters. Using 4-Methylumbelliferyl butyrate as a substrate we measured the inhibitory activity of JCP251 against rFphB and found that it has a fast inactivation rate Kobs/[I] of 8.5 ± 1.6 *106 [M−1 s−1] consistent with its ability to block labeling of FphB even at nanomolar concentrations in the competition labeling studies.
Fig. 3. Biochemical characterization of FphB and development of an FphB-selective fluorescent ABP.
a) Processing of 4-methylumbelliferyl-based fluorogenic substrates by rFphB. Turnover rates for each substrate are depicted as relative fluorescence units/min. Graph shows individual values overlaid with the means ± SD of 3 biologically independent samples. b) Proteolytic activity of rFphB or Trypsin against FITC-casein depicted as normalized fluorescence units. Graph shows an overlay of individual values and means ± SD, n= 3 biologically independent samples. Raw data were normalized by subtracting background fluorescence under buffer control conditions. c) Chemical structure of the fluorescent ABP JCP251-bT. d) SDS-PAGE analysis (TMR-fluorescence scan) of S. aureus Newman transposon mutant strains with insertions in indicated genes labeled with 100 nM JCP251-bT during exponential growth. (Full gel image in Fig. S4b) e) SDS-PAGE analysis of indicated Gram-positive and Gram-negative bacterial pathogens labeled with FP-TMR or JCP251-bT. MR-SA: S. aureus USA300, MS-SA: S. aureus ATCC35556, S.e.: S. epidermidis, L.m.: Listeria monocytogenes, E.c.: Escherichia coli, S.t.: Salmonella typhimurium, H.p.: Helicobacter pylori. Arrow indicates FphB. All experiments were repeated twice with similar results.
A fluorescent activity-based probe for S. aureus FphB
JCP251 is an ideal starting point for the design of an ABP for imaging of FphB activity in live bacteria. Our set of screening hits indicated that substitution of the C3 methoxy group by a propyloxy or isopropyloxy moiety (compound 3 (JCP174), compound 4 (JCP222) in Figure S1) largely retained the activity profile of JCP251. We therefore tested an analog of JCP251 containing the bodipyTMR fluorophore attached through a linker at the C3 methoxy substitution (compound 5 or JCP251-bT; Fig. 3c)38. This fluorescent probe had similar potency compared to the parent JCP251 inhibitor with only a ~3-fold lower inactivation rate (Kobs/[I] = 2.6 ± 1.8 *106 [M−1 s−1]). When JCP251-bT was applied to S. aureus cultures, it labeled a single protein that was observed throughout exponential and stationary phase growth in liquid culture (Supplementary Fig. 4a). Importantly, this specific labeling was lost in the FphB-transposon mutant strain, confirming the high selectivity of the probe for FphB in live cells (Fig. 3d, Supplementary Fig. 4b). Interestingly, the probe labeled a small number of potential off targets when applied to the soluble fraction of bacterial lysates instead of live cells (Supplementary Fig. 4c). This result could be due to limited membrane permeability of the probe or the release and activation of hydrolase targets under lysis conditions.
In order to further assess the selectivity of JCP251-bT for S. aureus FphB over related hydrolases in other bacteria, we used the probe to label various Gram-positive and Gram-negative bacteria (Fig. 3e). All of these bacteria express multiple serine hydrolase activities that were detected by the broad-spectrum FP-TMR probe. We performed homology searches for FphB and found orthologs with 53–57% identity in other Staphylococcal species, while putative orthologs in other prokaryotic and eukaryotic organisms showed only 25–39% identity (Supplementary Fig. 5a). Accordingly, JCP251-bT labeled methicillin-sensitive and methicillin-resistant S. aureus strains, as well as S. epidermidis, but did not show any labeling of targets in a number of other bacterial pathogens. To assess if JCP251-bT is sufficiently selective for FphB to be used for imaging during an infection, we performed labeling studies of live bacteria co-cultured with peripheral mononuclear cells and RAW cells. The probe weakly labeled proteins in both host cell types alone, but upon co-incubation of these cells with S. aureus, it predominantly labeled FphB (Supplementary Fig. 5c) and allowed visualization of probe-labeled bacteria over weak background fluorescence caused by non-specific probe uptake and/or labeling of host cell proteins (Supplementary Fig. 5d).
FphB activity is dynamically regulated
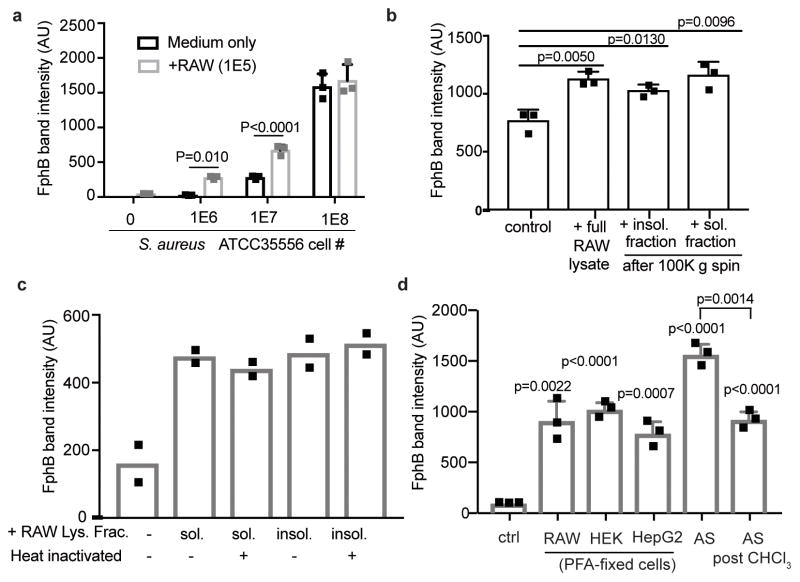
Given that expression levels of the fphB gene may be altered by skin derived signals37, we wanted to determine if activity levels (as measured by the JCP251-bT probe) responded to host cell derived factors. Interestingly, we observed that FphB labeling by JCP251-bT is significantly increased in the presence of RAW cells and that the extent of this effect was dependent on the ratio of bacterial cells to host cells (Fig. 4a and Supplementary Fig. 6a). This ratio-dependency might be explained by a contact-dependent mechanism where at high bacterial density the necessary contact points on the macrophage surface are saturated resulting in a lower net stimulation per bacterium. We observed that this stimulatory effect is retained in both soluble and insoluble fractions from RAW cell lysates as well as in PFA-fixed cells (Fig. 4b,c Supplementary Fig. 6b-e). Our data suggest that the increase in FphB activity is not dependent on active metabolic processes in the host cell, but is rather mediated through contact with a heat-stable component on the RAW cell surface. Notably, we did not observe any secretion of FphB into the culture medium (Supplementary Fig. 6f). Furthermore, the effect is not specific for macrophages. Bacteria reacted similarly to exposure to PFA-fixed HepG2 and PFA-fixed HEK cells, as well as to Fetalplex™ Animal Serum Complex. Notably, the stimulatory components in the serum could partially be depleted by chloroform-extraction suggesting they may be lipids or fatty acids. (Fig. 4d, Supplementary Fig. 6g) Together, these data suggest that S. aureus (both strains ATCC35556 and Newman) regulates FphB activity in response to host cell derived signals.
Fig. 4. Stimulatory activity of eukaryotic cell components on FphB activity.
All graphs show FphB activities in S. aureus cells in response to the indicated stimuli. After stimulation active FphB in live cells was labeled with JCP251-bT (100 nM), before lysates were analyzed by SDS-PAGE and FphB band intensities were quantified using Image Studio Lite. a) FphB band intensity in S. aureus ATCC35556 grown individually or in co-culture with RAW cells at the indicated cell numbers. Graph shows means ± SD of 3 biologically independent samples, p-values were calculated by unpaired, two-tailed Student’s t-test. See Fig. S6a for full gel. b) FphB band intensity after S. aureus ATCC35556 (1E7 CFU) were grown in the presence of full RAW cell lysate, or the insoluble and soluble fractions after ultracentrifugation (each equivalent to1E5 cells) prior to JCP251-bT labeling (see Fig. S6b for full gel). Graph shows means ± SD of 3 biologically independent samples, p-values were calculated by unpaired, two-tailed Student’s t-test. c) FphB band intensity after S. aureus ATCC35556 were treated with heat-inactivated or untreated soluble/insoluble lysate fractions after ultracentrifugation prior to JCP251-bT labeling. Graph shows means of 2 biologically independent samples. See Fig. S6d for full gel. d) FphB band intensity after S. aureus Newman WT or fphB::ϕNΣ (fphB) were incubated with PFA-fixed RAW, HEK, or HepG2 cells, 10% fetal animalplex serum or 10% fetal animalplex serum after chloroform extraction prior to labeling with JCP251-bT. See Fig. S6g for full gel. Graph shows means ± SD of three biologically independent samples, p-values were calculated by unpaired, two-tailed Student’s t-test.
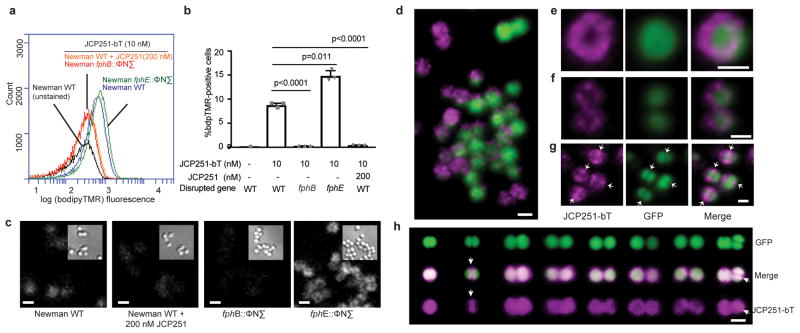
FphB labeling in the cell envelope is asymmetric and heterogenous
As the location of a protein provides clues to its physiological function, we sought to experimentally determine the location of enzymatically active FphB in the bacterium by labeling with JCP251-bT. To confirm probe specificity, we used flow cytometry to quantify labeling on a population scale in both WT and fphB transposon mutant bacteria (Fig. 5a,b, Supplementary Fig. 7). JCP251-bT labeling was detected in Newman WT and FphE-mutant populations, while for both the FphB-deficient bacteria and WT bacteria pretreated with JCP251 we only could detect background autofluorescence. Interestingly, we observed that the probe labeled only ~7–10% of Newman WT and a higher percentage of cells were probe-positive in the FphE-deficient strain. This suggests that FphB is heterogeneously expressed among individual cells within the bacterial population. Confocal fluorescence microscopy experiments in Newman cells again confirmed that JCP251-bT labeling is specific to FphB (Fig. 5c). To better localize FphB activity, we probe-labeled a Newman strain expressing a GFP reporter. We observed that FphB activity levels vary dramatically among individual WT cells (Fig. 5d) and as expected, pretreatment with JCP251 reduced fluorescent signals to background levels (Supplementary Fig. 8). Importantly, JCP251-bT labeling did not co-localize with the cytosolic GFP signal but rather showed concentrated signals in specific regions of the cell envelope (Fig. 5d–g). Furthermore, a subset of dividing cells showed enriched probe labeling in the septal cross-wall, the primary location of cell wall biogenesis in S. aureus (Fig. 5g,h) suggesting that FphB might modify substrates located in the bacterial cell wall.
Fig. 5. Imaging of FphB-activity using the fluorescent ABP JCP251-bT.
a) Flow cytometry plot of levels of JCP251-bT probe fluorescence in S. aureus Newman WT, fphB:: ϕNΣ and fphE:: ϕNΣ strains treated with FphB-inhibitor JCP251 or vehicle prior to labeling with JCP251-bT during exponential growth. b) Plot of percentage of cells within the BT-positive gate (see gating strategy in Fig. S8). Graph shows means ± SD of 3 biologically independent samples, indicated p-values were calculated by unpaired, two-tailed Student’s t-test. c) Confocal micrographs of indicated S. aureus strains labeled with 10 nM JCP251-bT. bT-fluorescence is depicted in white, insets show differential interference contrast (DIC) images. d) 3-d reconstruction of a series of confocal images of S. aureus Newman-GFP labelled with 10 nM JCP251-bT. GFP-fluorescence: green, bT-fluorescence: purple. e,f) Confocal micrographs of S. aureus Newman-GFP cell labeled with 10 nM JCP251-bT during exponential phase. g) Confocal micrograph of dividing S. aureus Newman-GFP labelled with 10 nM JCP251-bT during stationary phase. Arrows indicate division septum plane. h) 3-d reconstructions of confocal image series of S. aureus Newman-GFP cells labeled with 10 nM JCP251-bT during stationary phase. Examples of cells in different stages of cell division are shown. Enriched JCP251-bT labeling of the division septum is marked by white arrows. All scale bars: 1 μm. All experiments were repeated twice with similar results.
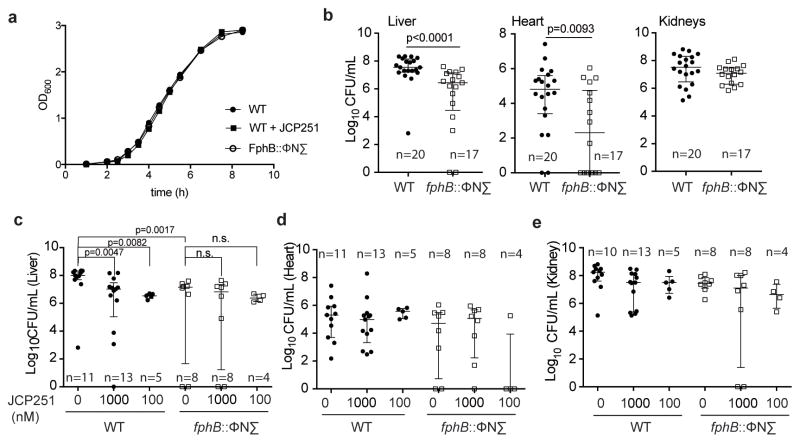
FphB is important for infection of distinct tissue sites in vivo
Given that FphB activity is concentrated in the cross-wall of dividing cells, we reasoned that it might be important for cell division. We therefore monitored the fitness of the fphB transposon mutant during in vitro growth. Surprisingly, the FphB-deficient strain had no growth defect in liquid culture (Fig. 6a) and showed a similar doubling time (55.4 ± 3.5 min) to the WT (55.9 min ± 1.5 min). Furthermore, treatment of WT bacteria with JCP251 to chemically block FphB activity did not alter bacterial growth, either (Fig. 6a). Given these results, and the fact that FphB activity levels are dynamic and change in response to host cell-derived factors, we reasoned that it may be functionally important during infection of a host organism. Using a systemic murine infection model, we observed that mice infected with the FphB transposon mutant strain had ~10–100 fold reduced bacterial loads compared to the WT strain in liver and heart tissues, but equal CFU numbers in the kidneys (Fig. 6b). In contrast, infection with the FphE transposon mutant only resulted in a small, but statistically significant reduction of bacterial loads in the liver, but not in the hearts or kidneys of infected mice (Supplementary Fig. 9). The overall weight loss in the course of infection with either mutant strain was comparable to that of WT-infected mice (Supplementary Fig. 9). These findings suggest the existence of distinct molecular mechanisms for the colonization of these organs by S. aureus and establish that FphB is important for S. aureus to infect heart and liver tissues.
Fig. 6. Effects of loss of FphB activity on infectivity in vivo.
a) Growth curves of S. aureus Newman and its isogenic transposon mutant strain fphB:: ϕNΣ and Newman WT in TSB with or without 1 μM JCP251, as indicated. Graph shows means of three (WT) or two biologically independent cultures (WT +JCP251, fphB:: ϕNΣ). b) Plots of total CFU recovered from indicated organs of BALB/c mice 96 hours after intravenous infection with S. aureus Newman WT or fphB:: ϕNΣ. Graphs show pooled data (median ± interquartile range, WT: n=20, fphB:: ϕNΣ. n=20) from 4 independent experiments. Significance was tested by unpaired, two-tailed Mann-Whitney test. c–e) Plots of total CFU recovered from indicated organs of BALB/c mice 96 hours after systemic infection with S. aureus Newman WT and fphB:: ϕNΣ that were pretreated with JCP251 or vehicle for 60 min. Graph shows pooled data (median ± interquartile range, sample size of different groups is indicated in the graph) from 2 independent experiments. Kruskal-Wallis test of analysis of variance among each of these datasets revealed that only in the liver median CFU values vary significantly (liver: p=0.0058, heart: p=0.1530, kidneys p=0.0956). Pairwise significance testing within the liver dataset (as indicated in c) was done by unpaired, two-tailed Mann-Whitney test.
Finally, we wanted to determine if chemical inhibition of FphB prior to infection would negatively affect the pathogen’s ability to establish tissue infection in vivo. We reasoned that pretreating cells with JCP251 would ‘chemically knock out’ FphB activity for early events during infection, but would eventually be overcome by de novo expression of the protein in vivo. Indeed, pretreatment of S. aureus Newman with JCP251 before infection reduced the bacterial burden in the liver to levels comparable to those observed upon infection with the fphB transposon mutant strain (Fig. 6c). In contrast, chemical knock-down of FphB prior to infection was insufficient to reduce CFU levels in the heart or kidneys (Fig. 6d,e). These data suggest a role for FphB activity during the early colonization/attachment phase of an intravenous infection.
Discussion
Although it is clear that serine hydrolases play important roles in various aspects of both bacterial and host cell biology, surprisingly little is known about this family of enzymes in S. aureus. This report describes the use of the serine-reactive electrophilic probe FP-TMR to identify putative serine hydrolase targets in S. aureus. Our profiling data confirm that S. aureus expresses a number of active serine hydrolases during normal and biofilm-like growth (in liquid culture and on agar) the majority of which are poorly characterized, lack annotation and have few, if any, mammalian homologs. The fact that this class of enzymes has so far largely escaped identification in ingg vitro phenotypic screens might be due to functional redundancy or functions in the host-pathogen interface that are difficult to capture with in vitro model systems.
Functional studies of one of these newly identified hydrolases, FphB, suggest that it responds to non-proteinaceous host-derived factors, is localized to the cell surface as well as the division septum and plays important roles in the early stages of infection in vivo. The finding that disruption of FphB expression results in defects in infection of the heart and liver but not the kidneys suggests FphB may help bacteria to functionally adapt during colonization. The observed differences in infectivity for specific target organs could be linked to a molecular surface architecture that may require different strategies for attachment. Glycosaminoglycans are common receptors for bacterial attachment or other aspects of bacterial pathogenesis39. Murine kidney cells have been shown to have higher levels of glycosaminoglycans such as heparan sulfate compared to liver and heart tissues40 and might thus facilitate FphB-independent tissue attachment. If FphB is in fact a key regulator of initial contact of bacteria with host cells, it could represent an ideal target for small molecules that could prevent the spread of a primary infection to other sites that cause increased morbidity and mortality (i.e. heart valves and artificial implants).
Although active during all culture stages and conditions tested, we found increased levels of FphB activity after contact with structural components of eukaryotic cell membranes and lipophilic serum components. Such contact is likely to occur upon initial attachment with host cells during early stages of an infection, where FphB activity could promote the establishment of infection. Thus, after sensing a molecular environment corresponding to a certain niche, elevated FphB activity might represent one element of a dynamic functional response of the bacteria to colonize or survive in this new environment. As we observed FphB activity only in a subset of cells, our results suggest that S. aureus populations display a degree of functional diversity that might facilitate their survival in a variety of biological niches.
How FphB mediates such effects and what its physiological substrates are, remains speculative. Our substrate profiling experiments with recombinant FphB suggest that the enzyme acts as a carboxylic acid esterase able to process C4-C8 chain fatty acid esters. Yet, the true physiologic substrate(s) may not have been accounted for in the limited set of synthetic fluorogenic substrates tested. As FphB activity is increased in the presence of host-derived components, one possible scenario is that the enzyme processes exogenous biomolecules allowing bacteria to scavenge host-derived substrates for their own metabolism. The ability to metabolize certain substrates may be key for survival in certain biological niches during infection or colonization (e.g. heart and liver), but may not affect growth in rich liquid culture media or other tissue sites (e.g. kidneys), consistent with our observations of the FphB mutant.
The potential endogenous substrate space includes the bacterial membrane and cell wall. As we did not observe cleavage of long chain fatty acid esters, we do not expect FphB to act on canonical membrane lipids. A first indication that FphB might directly act on substrates localized in the cell wall, is the observed enrichment of JCP251-bT labeling in the cross-wall of dividing cells. The fact that several studies found fphB gene expression moderately upregulated in response to cell wall stress conditions induced by the antibiotics oxacillin31 and daptomycin34, cationic antimicrobial peptides32, or the lantibiotic mersacidin33 further links FphB activity to this cellular location. Enzymes known to act in the cross-wall of dividing cells are important for shaping the cell wall as a structural scaffold protecting the inside of the bacterial cell (e.g. murein hydrolases)7 or as an outward-facing matrix for interaction with the environment (e.g. the Sortase A)41. Murein hydrolases are crucial for cell wall integrity and cell division and mutations in their corresponding genes invoke severe growth phenotypes in vitro7. We did not observe a similar phenotype for the fphB mutant. In contrast, Sortase A anchors secreted proteins with an LPXTG consensus sequence (e.g. adhesins, protein A and further virulence factors) to the free amino-terminus of pentaglycine in nascent peptidoglycan and deficiency in Sortase A attenuates pathogenicity in vivo42. Thus, Sortase A is a prime example of a cell surface-modifying enzyme whose functional relevance becomes evident in vivo, but not during growth in liquid culture. Similarly, FphB activity might govern environmental interactions by acting on cell-wall anchored proteins or structural components of the cell wall. Structural modifications of cell wall polymers that have been been reported to affect virulence, pathogenesis or colonization include O-acetylation at C6 of N-acetylmuramic acid in peptidoglycan43,44 and glycosylation45 and D-Alanylation46 of wall teichoic acid. While biosynthetic routes for the major cell envelope polymers are mostly well understood, evidence of enzymes involved in their dynamic modification or degradation is only emerging. Regulation of O-acetylation levels by the O-acetylpetpidoglycan esterase Ape, a serine hydrolase, has been described in different Gram-negative and Gram-positive bacteria, but not S. aureus47,48, suggesting this function may be inherent to a previously uncharacterized hydrolase in this organism. A recent study on the S. aureus methicillin-resistance factor FmtA demonstrated that this enzyme acts as a teichoic acid-specific D-Ala esterase49.
Interestingly, so-called Functional Membrane Microdomains (FMMs) are also associated with septal invaginations of dividing cells. These dynamic membrane-domains are organized through the scaffold protein flotillin, are largely composed of saccharolipids derived from the carotenoid staphyloxanthin, and their integrity is required for the activity of the cell-wall acting PBP2a, a mediator of penicillin-resistance in MRSA50. Although FphB was not found among FMM-associated proteins in MRSA50, it is conceivable that this enzyme acts on membrane-associated substrates in a regulatory fashion that translates into altered cell wall architecture with downstream effects on virulence. Given the multitude of described effects that individual cell envelope components have on bacterial virulence, regulated, dynamic chemical modifications of their structure through hydrolytic enzymes such as FphB appears to be a plausible strategy for bacteria to functionally adapt to their environment during infection or colonization.
The identification of FphB and the further set of 9 uncharacterized Fph serine hydrolases by ABPP provides a starting point for future elucidation of their corresponding biological functions. Our study demonstrates that these enzymes are ideal targets for the development of selective inhibitors and activity-based probes that will facilitate the characterization of these hydrolase activities in their physiological environments. Given these initial functional studies of FphB, inhibition of this target may represent a new targeted strategy to prevent S. aureus colonization in major organs, for example for prophylactic treatment of MRSA-carriers before surgery. Furthermore, the ability to selectively label FphB with a small molecule fluorescent probe renders this enzyme a potential target for the development of targeted probes for localization and risk stratification of Staphylococcus spp. infections. These types of therapy and imaging agents could have a broad impact on diagnosis, treatment and therapy response monitoring for diverse types of S. aureus infections.
Online methods
Bacterial strains and growth conditions
All bacterial strains employed in this study are summarized in Table S4. S. aureus strains ATCC3555, USA300, Newman or its isogenic mutants were used as indicated. S. aureus and S. epidermidis were routinely cultured in Difco tryptic soy broth (TSB; BD, Sparks, MD), or on tryptic soy agar (TSA; BD). For compound screening and hit validation, strain ATCC3556 was grown on TSAMg (Tryptic Soy Agar with 100 mM MgCl2) for 48–72 h. When required to select for transductants, erythromycin and lincomycin were added to the media at concentrations of 3 μg/mL and 20 μg/mL, respectively (Sigma Aldrich, St. Louis, MO). All incubations were performed at 37°C, and liquid cultures were aerated by shaking at 180 rpm unless indicated otherwise. The GFP plasmid pCM2951 was obtained from Dr. Alexander Horswill (University of Iowa) and transformed into WT S. aureus Newman.
For growth curves of S. aureus Newman and its fphB transposon mutant, overnight cultures were diluted 1:400 in 30 mL fresh TSB and grown in 125 mL culture flasks at 180 rpm, 37°C. Culture media included 1 μM JCP251 or the same volume (0.1%) of vehicle (DMSO) under control conditions. Culture aliquots were taken in 30–60 min intervals, diluted with TSB (1:1 – 1:3) dependent on the growth phase and OD600 values were determined in a Laxco DSM-Micro Cell Densitometer.
S. typhimurium and E. coli were grown in Luria Bertani (LB) broth or on LB agar. L. monocytogenes was grown in BHI broth or on BHI agar. H. pylori was grown in Brucella broth or on Columbia agar.
Cell lines and purification of peripheral mononuclear cells
RAW cells and HEK 293 cells were grown in Dulbecco’s Modified Eagle Medium (supplemented with 10% Fetalplex™ Animal Serum Complex (Gemini Bio-Products), 2 mM L-glutamine and 1% penicillin/streptomycin). HepG2 cells were grown in DMEM with low glucose, L-glutamine, sodium pyruvate (Caisson Labs DML28), supplemented with 10% Fetalplex™ Animal Serum Complex and 1% penicillin/streptomycin. Cells were grown to 90% confluency, washed 1x with phosphate buffered saline (PBS) and harvested with a cell lifter. Cell pellets were centrifuged at 1,200 rpm, 3 min and taken up in the desired culture medium. Whole human blood samples were received from the Stanford Blood Bank and peripheral mononuclear cells (PMNs) were purified using the EasySep™ Direct Human Neutrophil Isolation Kit (StemCell Technologies).
Genetic manipulation of S. aureus strain Newman
Oligonucleotides used in confirming successful transduction of serine hydrolase mutants into S. aureus Newman can be found in Table S5. In brief, transposon mutagenesis was used to generate insertion mutations in each NWMN_0169, NWMN_0262, NWMN_0748, NWMN_1210, NWMN_1683, NWMN_2092, NWMN_2350, NWMN_2379, NWMN_2434, NWMN_2480, NWMN_2528, and NWMN_2569. Homologs of the aforementioned genes were first identified through the Basic Local Alignment Search Tool (BLASTn) in S. aureus USA300 FPR3757 (GeneBank ID CP000255.1), and corresponding transposon mutants were identified and confirmed in the Nebraska Transposon Mutant Library30 through PCR. The mutations were mobilized into S. aureus Newman using transduction with phage ϕ85. Successful transductants were confirmed through PCR and assessed for hemolysis relative to wild-type S. aureus Newman and USA300 by streaking on BD Trypticase Soy Agar II with 5% Sheep’s Blood (BD).
FphB sequence analyses
The FASTA sequence of Q2FV90 (SAOUHSC_02844) was retrieved from Uniprot and analyzed bioinformatically using the following webtools with settings for Gram-positive bacteria, if applicable: Secretome P2.0 (http://www.cbs.dtu.dk/services/SecretomeP/), PSORTB Ver 3.0.2 (http://www.psort.org/psortb/) Signal P4.1 (www.cbs.dtu.dk/services/SignalP/), Signal-3L 2.0 and MemBrain (http://www.csbio.sjtu.edu.cn) and TmPred (minimum length: 14, maximum: 33) (http://www.ch.embnet.org/software/TMPRED_form.html).
Phylogenetic analysis
The FphB protein sequence (Uniprot ID: Q2FV90, Strain NCTC8325, gene SAOUHSC02844) was used as a blastp (protein-protein blast, https://blast.ncbi.nlm.nih.gov) query sequence in a homology search against the non-redundant protein sequences database for different organisms. Top hits obtained for each organisms were subjected to a Clustal Omega Multiple Sequence Alignment (http://www.ebi.ac.uk/Tools/msa/clustalo/) and the phylogenetic tree was visualized using iTOL:interactive Tree Of Life (itol.embl.de).
Cloning of SAOUHSC02844 (fphB) gene
Genomic DNA was purified from S. aureus ATCC35556 using the Qiagen DNEasy Blood & Tissue kit (Qiagen, Hilden, Germany) and used as a template for PCR-amplification of the SAOUHSC_02844 gene using primers SAOUHSC02844_Fw_BamHI and SAOUHSC02844_Rv_XhoI with BamHI and XhoI restriction sites (Table S5). The construct was cloned into the pET28a vector.
Expression, purification and labelling of rFphB
The pet28a-FphB expression vector was transformed into chemically competent BL21 (DE3) E. coli. An overnight culture of the transformed bacteria in LB (+Kanamycin 35 μg/ml) selection medium was diluted 1:400 into 2 L selection medium (in a 4 L flask) at 37 °C, 220 rpm. At OD600 0.1 – 0.2 the culture was continued to grow at 27°C and protein expression was induced at OD600 of ~ 0.4 by addition of IPTG (10 μM final concentration). After growth for 3 h at 27°C, 220 rpm, cells were harvested by centrifugation, transferred to two 50 mL conical tubes, centrifuged again and bacterial pellets stored at −80°C.
Each pellet was thawed on ice, resuspended in 20 mL lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 1% TritonX-10, pH 8.0) and lysed by sonication (6x 10 s, 1.5 s pulse, 30% Amplitude, Branson Sonifier). Lysates were centrifuged at 4,500 g, 4 °C for 45 min and the supernatant transferred to a 15 mL conical and a 50% slurry of His60 Ni Superflow Resin (Clontech Laboratories) was added and incubated at room temperature (RT), rotating for 60 min, then purified by gravity flow. The resin was washed 3x with 20 mM NaH2PO4, 300 mM NaCl,20 mM imidazole, 1% TritonX-10, pH 8.0 and the His6- and T7-tagged protein was eluted in 50 mM NaH2PO4, 300 mM NaCl, 200 mM imidazole. Eluates were labeled with 1 μM FP-TMR and analyzed by SDS-PAGE (TMR-fluorescence scan and Coomassie stain). rFphB containing fractions were pooled and were further purified using the Novagen T7 Tag Affinity Purification Kit as per the manufacturer’s instruction (EMD Millipore, Billerica, MA). Finally, purified rFphB was dialyzed 2x into 50 mM Tris-HCl, 300 mM NaCl and glycerol was added to a final concentration of 10% for long-term storage at −80°C. The concentration of purified rFphB was determined by A280 measurements using a calculated E1% of 10.76.
For ABP-labeling experiments, rFphB (50 nM) in PBS/0.01% SDS was pre-incubated with different concentrations of JCP251 or DMSO for 30 min at 37 °C, before JCP251-bdpTMR (100 nM) or FP-TMR (1 μM) were added and the sample mixed by pipetting. Samples were incubated at for 37 °C for 30 min, boiled in 1xSDS-Loading buffer and analyzed by SDS-PAGE.
Bacterial labeling with fluorescent ABPs
After growth on agar plate or in liquid culture as indicated, bacteria were suspended to the desired density in TSB and added to microtubes in a final volume of 50 – 300 μL. For inhibitor pretreatment experiments, the inhibitors were added from 100x-concentrated stock solutions in DMSO and incubated for 60 min (37 °C, 300 rpm) prior to ABP-labeling. FP-TMR (1 μM) or JCP251-bT (10 nM or 100 nM) were added from 50–100X stock solutions in DMSO and cells incubated for 30 min, at 37 °C, 300 rpm. Samples were then taken up in 1x SDS-Loading Buffer and lysed by bead beating as described below.
For comparative bacterial labeling experiments H. pylori and L. monocytogenes were grown in 5 mL of liquid culture. Cultures were spun down and cell densities adjusted to ~2E8 CFU/100 μL in TSB. All other bacterial strains were scraped off agar plates and resuspended in TSB at equivalent concentrations. To 100 μL aliquots of bacterial suspension was added 1 μL of 100 μM FP-TMR or 10 μM JCP251-bT. Cells were mixed by pipetting and incubated at 37 °C for 30 min. 1 mL PBS was added and samples were centrifuged (8,000 g, 5 min, RT). The supernatant was taken off and cell pellets resuspended in 500 μL 1x SDS-Loading Buffer and lysed by bead-beating.
Competitive activity-based protein profiling screen
Experimental details on the screening procedure are given in the ‘Small Moleule Screening Data Table’ (Table S1).
Bacterial lysis
After probe labeling bacterial suspensions were prepared PBS/0.1% SDS or already in 1xSDS-Loading Buffer. Suspensions were transferred to 0.6 mL (400–500 μL samples) or 2.0 mL (1.0 – 1.2 mL samples) o-ring tubes filled ~half with 0.1 mm glass beads (Biospec Products, Bartlesville, OK, USA). Tubes were placed in pre-chilled aluminum vial rack and samples lysed in a Mini-BeadBeater-96 (Biospec Prod) (3x 50 s with 2 min resting time on ice in-between) and centrifuged at 13,000g, 4 °C, 5 min.
SDS-PAGE analysis of samples labeled with fluorescent ABPs
Lysate supernatants were combined with 4X SDS-Loading Buffer. Samples were boiled at 95 °C for 10 min and separated by SDS-PAGE gel. TMR-fluorescence was scanned on a Typhoon 9410 variable mode imager (TAMRA channel, λex = 380, 580 BP filter).
Preparation of RAW cell lysates and fractionation
At ~90% confluency RAW cells were washed 2x in PBS scraped off plates and harvested in DMEM (+2 mM L-glutamine). Cells were spun down by centrifugation at 1,200 rpm, RT and cells adjusted to a concentration of 1E5 cells/50 μL. Cell suspensions were cooled on ice, transferred into 7 mL vials filled with ~ 3.5 mL of 0.1 mm glass beads and lysed by bead beating (1x 50 s). Supernatants were stored at −80°C or further fractionated by ultracentrifugation at 100,000 g, 60 min, 4 °C in a Optima™ MAX-TL Ultracentrifuge (Beckman Coulter, Indianapolis, IN, USA).
ABP labeling experiments in eukaryotic/bacterial cell co-cultures
Labeling to assess the selectivity of JCP251-bT
RAW cells or PMNs (~50,000) were seeded out in RPMI/HEPES into sterile flat-bottom 96-well plates. The plate was spun down at 1,200 rpm (accel=5, brake=3) for 3 min and incubated at 37°C, 5% CO2 for 60 min. S. aureus ATCC3556 was grown overnight on TSA, scraped off the plate and adjusted to an OD600 of 0.5 in DMEM (+2 mM L-glutamine). Eighty μL of bacterial suspension (1E7 CFU, MOI=200) or medium were added to either cell type or control wells containing medium only and incubated for 60–90 min at 37°C, 5% CO2, before JCP251-bdpTMR was added to a final concentration of 100 nM. Plates were spun down, incubated at 37°C, 5% CO2. 60 μL 4xSDS-Loading Buffer was added to lyse eukaryotic cells. Samples were transferred to o-ring-tubes for lysis and SDS-PAGE analysis.
FphB stimulation experiments
S. aureus ATCC35556 was harvested after growth on TSA or S. aureus Newman was grown to exponential phase in TSB, centrifuged. Bacteria were resuspended in serum- and antibiotic free culture medium (DMEM, 2 mM L-glutamine or RPMI, 5 mM HEPES) and incubated for 60–90 min in the presence of different stimulatory conditions prior to labeling with 50 – 100 nM JCP251- bdpTMR. Stimulatory conditions include: RAW cells or PMNs seeded in serum- and antibiotic free culture medium, fixed RAW, HEK 293 or HepG2 cells (treated with 2% PFA for 10 min and washed 3x with PBS), Fetalplex™ Serum (before and after extraction with an equal volume of chloroform) full RAW cell lysate or soluble and insoluble fractions after ultracentrifugation, as well as heat-inactivated lysate fractions. Heat-inactivation of lysate fractions was done at 95°C for 15 min. After probe labeling, cell lysis and SDS-PAGE analysis FphB bands were quantified using Image Studio Lite software (LI-COR Biosciences). Statistical analysis was done using Prism 7.0 (GraphPad Software)
Fluorogenic substrate assays
Stock solutions of 4-Methylumbelliferyl(4-MU)-based fluorogenic substrates were dissolved in DMSO (1 mM) and 0.3 μL were added to the wells of an opaque flat-bottom 384 well plate. Thirty μL of a solution rFphB in PBS/0.01%TritonX-100 (400 pM active enzyme) was added and fluorescence (λex = 365 nm and λem = 455 nm) was read at 37 °C in 1 min intervals on a Cytation 3 imaging reader (BioTek, Winooski, VT, USA) for 60 min. Turnover rates in the linear phase of the reaction (10 – 40 min) were calculated using Gen5 software (BioTek) as RFU/min. Rates were normalized by subtracting background hydrolysis rates measured for each substrate in reaction buffer in the absence of protein.
Determination of inactivation constants kobs/[I]
Inactivation rates kobs/[I] of recombinant FphB by inhibitors JCP251 and JCP251-bT were measured under pseudo first-order conditions at a substrate concentration of 1 mM 4-methylumbelliferyl butyrate (4-MU butyrate). To 0.3 μL substrate (in DMSO) were added 0.3 μL of inhibitor (in DMSO) to the wells of a 384-well plate and, after addition of 29.6 μL FphB (0.8 nM active enzyme) in PBS/0.01%TritonX-100, the enzymatic reaction was monitored as described above at 37 °C for 30 min. Data were normalized by subtracting background signal due to substrate hydrolysis (in the absence of FphB) at each timepoint. The normalized progress curve was fit to the formula P = V/kapp *(1 − e kapp*t) + C with P = product formation, V = initial velocity, t= time and kapp as the apparent rate constant. A constraint was set for V as the rate determined for the linear, uninihibited reaction for each experiment. The second order rate constant kobs/[I] was then calculated using the formula kobs/[I] =kapp/[I] (1 + [S]/KM) for three different inhibitor concentrations (5 nM, 10 nM and 20 nM). The KM-value for this substrate was experimentally determined as 52 ±11 μM. Mean kobs/[I] values ± SD were determined from 3 independent experiments.
FITC-casein assay
A stock solution of FITC-casein (5 mg/mL in water) was diluted 1:500 in assay buffer (PBS/0.01%Triton X-100) and 50 μL added to the wells of an opaque flat-bottom 96-well plate. Fifty μL FphB (2 nM active enzyme), Trypsin (2 nM) or assay buffer were added and the plate incubated at 37 °C. After 30 min and overnight incubation fluorescein fluorescence (λex = 485 nm and λem = 538 nm) were measured on a Cytation 3 imaging reader.
FP-biotin labeling of S. aureus serine hydrolases and sample preparation for mass-spectrometry
S. aureus ATCC35556 was grown on TSBMg for 72 h and resuspended to an OD600 ~100 in 13 mL TSB. 1 mL aliquots were transferred to a 1.5 mL tube and JCP251 (300 nM, 1 μM), JCP678 (1 μM) or DMSO (5 μL) were added and cells incubated for 60 min at 37 °C, 700 rpm. Five μL of FP-biotin (1 mM in DMSO) were added to the bacteria and incubated for 30 min, before samples were spun down (4,500 g, 5 min, 4 °C) and the supernatant was aspirated. The cell pellet was resuspended in 1.2 mL PBS/0.1%SDS and 100 μL Protease Inhibitor Cocktail were added. The samples were transferred to 2.0 mL o-ring tubes half-filled with 0.1 mm glass beads, tubes placed into pre-chilled aluminum blocks and lysed by bead-beating (3x 50 s with 2 min on ice in-between) as described above. Samples were centrifuged for 5 min at 10,000 g, 4 °C. Fifteen μL aliquots were taken for SDS-PAGE analysis and SA-HRP blot. Protein concentration in the supernatant was adjusted to 0.8 mg/ml and 200 μL 10% TritonX-100 added to 0.8 mg of proteome and incubated at 4°C for 1 hr, rotating. Sample volume was brought to 2.5 mL and added to a pre-equilibrated PD10 desalting column (GE LifeSciences) discarding the flow through. Samples were eluted with 3.5 mL PBS and 10% SDS/PBS was added to a final concentration of 0.5% SDS. Samples were boiled at 90 °C for 8 min, then cooled in a −20°C freezer for ~ 5 min before ~80 μL of a prewashed 50% Streptavidin-agarose slurry was added and samples were incubated at RT for 1 hr, rotating. Samples were centrifuged at 300g for 2 min and washed 2x with 1% SDS, 2x with 6M urea and 2x with PBS (each 10 ml). After the final wash, resin was resuspended in 500 μL 6 M urea, 25 μL of DTT (30 mg/ml solution in PBS) and heated at 65 °C for 15 min. 25 μL of iodoacetamide (14 mg/mL in PBS) were added and incubated at 37 °C for 30 min. Following incubation, 950 μL PBS was added, the resin spun down and the supernatant discarded. A premixed solution of 200 μL of 2 M urea/PBS, 2 uL 100 mM CaCl2 in water and 4 μL Trypsin solution (20 μg lyophilized powder reconstituted in 40 μL 2 M urea/PBS) was added to the resin and samples incubated at 37°C overnight, rotating. Tryptic peptide digests were separated from the resin over a spin-filter column and resin washed 2x with 50 μL PBS. Fifteen μL of formic acid was added to the combined eluates for each sample. Samples were stored at −20°C until LC/LC-MS/MS analysis.
Liquid chromatography-mass spectrometry analysis
LC-MS/MS analysis was performed on an LTQ-Orbitrap Discovery mass spectrometer (ThermoFisher) coupled to an Agilent 1200 series HPLC. Peptide digests were pressure loaded onto a 250 μm fused silica desalting column packed with 4 cm of Aqua C18 reverse phase resin (Phenomenex). The peptides were eluted onto a biphasic column (100 μm fused silica with a 5 μm tip, packed with 10 cm C18 and 4 cm Partisphere strong cation exchange resin (SCX, Whatman) using a gradient of 5–100% Buffer B in Buffer A (Buffer A: 95% water, 5% acetonitrile, 0.1% formic acid; Buffer B: 20% water, 80% acetonitrile, 0.1% formic acid). The peptides were then eluted from the SCX onto the C18 resin and into the mass spectrometer using 4 salt steps previously described. The flow rate through the column was set to ~0.25 μL/min and the spray voltage was set to 2.75 kV. One full MS scan (FTMS) (400–1800 MW) was followed by 8 data dependent scans (ITMS) of the nth most intense ions.
The tandem MS data were searched using the SEQUEST algorithm using a concatenated target/decoy variant of the NCTC8325 S. aureus UniProt database. A static modification of +57.02146 on cysteine was specified to account for alkylation by iodoacetamide. SEQUEST output files were filtered using DTASelect. Proteins with less than 5 average spectral counts among the three replicates of the control sample were filtered from the dataset. Fold change was calculated as the quotient of the number of average spectral counts for the control sample divided by the number of average spectral counts for the compound treated sample. P values were calculated for the compound treated samples and control samples.
Flow cytometry
Labeled bacteria were were washed 2x in PBS, fixed in 2% PFA for 10 min and washed 3x with PBS. Bacteria were resuspended in PBS and analyzed on a BD Accuri™ RUO Special Order System Flow Cytometer with BD CSampler software (BD Biosciences, San Diego, USA). FSC-A threshold was set to 5,000 and 30,000 – 50,000 events in the bacterial cell gate were recorded. BdpTMR-fluorescence was detected using a 552 nm laser and 586 nm filter (FL2-detector).
Fluorescence microscopy
Imaging of RAW cells
1E5 RAW cells were seeded onto a sterile coverslip in a 24-well plate in 400 μL DMEM (+2.5 mM L-glutamine) and incubated at 37 °C, 5% CO2. The next day S. aureus ATCC3556 were harvested after growth on TSAMg and adjusted to an OD600 of ~ 0.8 in DMEM (+2.5 mM L-glutamine). Fifty μL (1E7 CFU) were added to the cells, incubated for 60 min at 37°C, 5% CO2, before 5 μL of 7 μM JCP251-bdpTMR (100 nM) was added and cells incubated for another 30 min at 37 °C, 5% CO2. After 30 min of labeling cells were washed 2x with PBS, fixed in 2%PFA for 10 min at RT and washed 3x with PBS. Coverslips were mounted onto microscope slides with VECTASHIELD Antifade Mounting Medium with DAPI (Vector Laboratories).
Imaging of bacteria only
A suspension of labeled and fixed bacteria in PBS was added onto an agarose pad on a microscope slide and covered with a coverslip. Cells were imaged at 63x using a Zeiss LSM 700 confocal microscope in the BodipyTMR and GFP channels. Laser and gain settings were configured at the beginning of each experiment and constant settings used for all samples.
Murine model of S. aureus systemic infection
Two days prior to infection, S. aureus Newman and its isogenic transposon mutants were streaked on TSA and incubated overnight at 37 °C. Single isolated colonies from the TSA plates were used to prepare overnight cultures in 5 mL of TSB. Overnight cultures were back-diluted 1:100 in 5 mL of TSB, and grown for 2.5 h to mid-exponential phase. Cultures were pelleted by centrifugation at 6,000 rpm for 10 minutes, and washed once in sterile PBS. Cells were normalized to an OD600 of 0.4 in PBS, corresponding to a cell density of approximately 1–2 x 108 CFU/mL. Each inoculum was quantified by serial dilution and plating on TSA.
For all murine experiments, female BALB/cJ mice were purchased from The Jackson Laboratory and infected at 6–8 weeks of age. In brief, mice were anesthetized by intraperitoneal injection with 125–250 mg/kg of 2,2,2-tribromoethanol and were subsequently infected with S. aureus through injection of 100 μL of prepared cells (~1–2 x 107 total CFU) into the retroorbital sinus. The infection was allowed to proceed for 96 h, with frequent monitoring throughout the course of the experiment.
Infection was allowed to proceed for 96 h, and the mice were assessed for health and weight every 12 and 24 h, respectively. Following euthanasia by CO2 inhalation, the kidneys, heart and liver were harvested. The extracted organs were homogenized and serially diluted in sterile 1 x PBS before plating for CFU on TSA. Plates were incubated overnight at 37°C and counted the following day. All murine experiments were evaluated and approved by the Institutional Animal Care and Use Committee (IUCAC) of Vanderbilt University Medical Center, and are in compliance with NIH guidelines, the Animal Welfare Act, and US Federal law.
In vivo inhibitor studies
To assess for the function of the JCP251 inhibitor in vivo, S. aureus wild-type and fphB::ϕNΣ cells were prepared as described above. Following washing and normalization in PBS, the inocula were mixed with 1 μM of the JCP251 inhibitor, and incubated with shaking at room temperature for 1 h. Mice were injected with 100 μL of the S. aureus-inhibitor mix, or with 100 μL of S. aureus in PBS with 1% DMSO, as a mock control for the inhibitor. The infection was allowed to proceed as above and bacterial burdens in the kidneys, liver and hearts were assessed after 96 h.
Statistical analyses
All statistical analyses were done using GraphPad Prism 7 (GraphPad Software Inc.). Details on sample size and statistical tests used are given in the corresponding figure legends. Unless stated otherwise data from in vitro experiments subject to statistical analysis was performed in 3 technical replicates and analyzed by unpaired Student’s t-Test. Representative data shown was performed at least in two independent experiments with a similar outcome. Data from S. aureus infection experiments were pooled from 2–3 different experiments. If nonparametric one way ANOVA (Kruskal Wallis test) indicated that median values of the different experimental groups within a dataset varied significantly, individual groups were compared by pairwise significance testing using unpaired, two-tailed Mann-Whitney-U test. P-values <0.05 were considered significant.
Extended Data
Extended Data Table 1.
| Protein | Mol Weight (Da) | JCP678_1uM_1 | JCP678_1uM_2 | JCP678_1uM_3 | ctrl_1 | ctrl_2 | ctrl_3 | JCP678_1uM_Average | ctrl_Average | p value | ctrl/678 | log ratio | log p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tr|Q2FUY3|Q2FUY3_STAA8 - Tributyrin esterase, putative OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_02962 PE=4 SV=1 xxxxx | 29096 | 8 | 75 | 47 | 520 | 244 | 125 | 43.33 | 296.33 | 0.10 | 6.84 | 2.77 | 3.32 |
| tr|Q2G0V7|Q2G0V7_STAA8 - Uncharacterized protein OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_00417 PE=4 SV=1 xxxxx | 27443 | 14 | 31 | 14 | 40 | 44 | 24 | 19.67 | 36.00 | 0.12 | 1.83 | 0.87 | 3.04 |
| tr|Q2G2D6|Q2G2D6_STAA8 - Uncharacterized protein OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_02448 PE=4 SV=1 xxxxx | 33186 | 5 | 5 | 6 | 12 | 12 | 5 | 5.33 | 9.67 | 0.14 | 1.81 | 0.86 | 2.84 |
| sp|Q2FV67|ROCA_STAA8 - 1-pyrroline-5-carboxylate dehydrogenase OS=Staphylococcus aureus (strain NCTC 8325) GN=rocA PE=3 SV=1 xxxxx | 56868 | 7 | 20 | 13 | 7 | 7 | 6 | 13.33 | 6.67 | 0.15 | 0.50 | −1.00 | 2.72 |
| sp|Q2FXL6|Y1819_STAA8 - Putative universal stress protein SAOUHSC_01819 OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_01819 PE=3 SV=1 xxxxx | 18475 | 8 | 17 | 10 | 6 | 9 | 5 | 11.67 | 6.67 | 0.17 | 0.57 | −0.81 | 2.57 |
| sp|Q2G028|ENO_STAA8 - Enolase OS=Staphylococcus aureus (strain NCTC 8325) GN=eno PE=1 SV=1 xxxxx | 47117 | 6 | 12 | 11 | 9 | 7 | 4 | 9.67 | 6.67 | 0.27 | 0.69 | −0.54 | 1.88 |
| tr|Q2FV90|Q2FV90_STAA8 - Uncharacterized protein OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_02844 PE=4 SV=1 xxxxx | 36848 | 60 | 277 | 68 | 323 | 264 | 137 | 135.00 | 241.33 | 0.30 | 1.79 | 0.84 | 1.73 |
| sp|Q2G0N1|EFG_STAA8 - Elongation factor G OS=Staphylococcus aureus (strain NCTC 8325) GN=fusA PE=1 SV=3 xxxxx | 76612 | 6 | 14 | 15 | 8 | 11 | 4 | 11.67 | 7.67 | 0.32 | 0.66 | −0.61 | 1.66 |
| sp|Q2G0Y7|IMDH_STAA8 - Inosine-5′-monophosphate dehydrogenase OS=Staphylococcus aureus (strain NCTC 8325) GN=guaB PE=3 SV=1 xxxxx | 52851 | 4 | 18 | 9 | 9 | 6 | 3 | 10.33 | 6.00 | 0.39 | 0.58 | −0.78 | 1.38 |
| tr|Q2G1C8|Q2G1C8_STAA8 - Uncharacterized protein OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_00197 PE=3 SV=1 xxxxx | 44729 | 19 | 60 | 28 | 33 | 28 | 12 | 35.67 | 24.33 | 0.46 | 0.68 | −0.55 | 1.11 |
| sp|P60430|RL2_STAA8 - 50S ribosomal protein L2 OS=Staphylococcus aureus (strain NCTC 8325) GN=rplB PE=1 SV=1 xxxxx | 30155 | 7 | 6 | 4 | 7 | 4 | 12 | 5.67 | 7.67 | 0.47 | 1.35 | 0.44 | 1.10 |
| tr|Q2FVA9|Q2FVA9_STAA8 - Uncharacterized protein OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_02824 PE=4 SV=1 xxxxx | 21793 | 12 | 24 | 6 | 14 | 10 | 4 | 14.00 | 9.33 | 0.48 | 0.67 | −0.58 | 1.05 |
| tr|Q2G1C9|Q2G1C9_STAA8 - Uncharacterized protein OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_00196 PE=3 SV=1 xxxxx | 84608 | 5 | 12 | 10 | 6 | 12 | 0 | 9.00 | 6.00 | 0.50 | 0.67 | −0.58 | 1.00 |
| sp|P0A0B7|AHPC_STAA8 - Alkyl hydroperoxide reductase subunit C OS=Staphylococcus aureus (strain NCTC 8325) GN=ahpC PE=1 SV=1 xxxxx | 20977 | 7 | 11 | 8 | 10 | 7 | 5 | 8.67 | 7.33 | 0.52 | 0.85 | −0.24 | 0.95 |
| sp|Q2FV39|Y2900_STAA8 - Uncharacterized hydrolase SAOUHSC_02900 OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_02900 PE=3 SV=1 xxxxx | 31006 | 118 | 384 | 276 | 409 | 257 | 293 | 259.33 | 319.67 | 0.54 | 1.23 | 0.30 | 0.89 |
| tr|Q2FVG3|Q2FVG3_STAA8 - Carboxylic ester hydrolase OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_02751 PE=3 SV=1 xxxxx | 51969 | 33 | 201 | 53 | 274 | 136 | 33 | 95.67 | 147.67 | 0.58 | 1.54 | 0.63 | 0.77 |
| tr|Q2G2C1|Q2G2C1_STAA8 - Pyruvate carboxylase OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_01064 PE=4 SV=1 xxxxx | 128548 | 812 | 2900 | 1198 | 1874 | 1079 | 786 | 1636.67 | 1246.33 | 0.62 | 0.76 | −0.39 | 0.70 |
| tr|Q2FYZ3|Q2FYZ3_STAA8 - Hydrolase, alpha/beta fold family domain protein OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_01279 PE=4 SV=1 xxxxx | 35256 | 24 | 111 | 30 | 89 | 62 | 52 | 55.00 | 67.67 | 0.70 | 1.23 | 0.30 | 0.52 |
| sp|P02976|SPA_STAA8 - Immunoglobulin G-binding protein A OS=Staphylococcus aureus (strain NCTC 8325) GN=spa PE=1 SV=3 xxxxx | 56437 | 58 | 186 | 142 | 287 | 123 | 66 | 128.67 | 158.67 | 0.71 | 1.23 | 0.30 | 0.49 |
| sp|Q2G2J2|SSAA2_STAA8 - Staphylococcal secretory antigen ssaA2 OS=Staphylococcus aureus (strain NCTC 8325) GN=ssaA2 PE=1 SV=1 xxxxx | 29327 | 4 | 16 | 12 | 14 | 7 | 6 | 10.67 | 9.00 | 0.72 | 0.84 | −0.25 | 0.47 |
| tr|Q2FY42|Q2FY42_STAA8 - Acetyl-CoA carboxylase, biotin carboxyl carrier protein OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_01624 PE=4 SV=1 xxxxx | 17122 | 22 | 169 | 73 | 92 | 66 | 58 | 88.00 | 72.00 | 0.74 | 0.82 | −0.29 | 0.44 |
| tr|Q2FXX0|Q2FXX0_STAA8 - Acetyl-CoA carboxylase, biotin carboxyl carrier protein, putative OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_01710 PE=4 SV=1 xxxxx | 16794 | 29 | 251 | 103 | 119 | 192 | 131 | 127.67 | 147.33 | 0.79 | 1.15 | 0.21 | 0.34 |
| sp|Q2FYU7|CATA_STAA8 - Catalase OS=Staphylococcus aureus (strain NCTC 8325) GN=katA PE=2 SV=2 xxxxx | 58380 | 3 | 10 | 4 | 4 | 7 | 4 | 5.67 | 5.00 | 0.80 | 0.88 | −0.18 | 0.33 |
| sp|Q2FUU5|LIP1_STAA8 - Lipase 1 OS=Staphylococcus aureus (strain NCTC 8325) GN=lipA PE=1 SV=1 xxxxx | 76675 | 0 | 21 | 9 | 11 | 8 | 16 | 10.00 | 11.67 | 0.81 | 1.17 | 0.22 | 0.30 |
| tr|Q2G2V6|Q2G2V6_STAA8 - Uncharacterized protein OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_01912 PE=4 SV=1 xxxxx | 28362 | 6 | 15 | 4 | 11 | 8 | 3 | 8.33 | 7.33 | 0.82 | 0.88 | −0.18 | 0.29 |
| tr|Q2G025|Q2G025_STAA8 - Carboxylesterase, putative OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_00802 PE=4 SV=1 xxxxx | 28094 | 19 | 247 | 91 | 153 | 91 | 61 | 119.00 | 101.67 | 0.82 | 0.85 | −0.23 | 0.28 |
| sp|Q2G0N0|EFTU_STAA8 - Elongation factor Tu OS=Staphylococcus aureus (strain NCTC 8325) GN=tuf PE=3 SV=1 xxxxx | 43104 | 13 | 47 | 24 | 24 | 33 | 21 | 28.00 | 26.00 | 0.86 | 0.93 | −0.11 | 0.22 |
| sp|Q2G155|LIP2_STAA8 - Lipase 2 OS=Staphylococcus aureus (strain NCTC 8325) GN=lip2 PE=1 SV=1 xxxxx | 76388 | 63 | 256 | 127 | 221 | 139 | 59 | 148.67 | 139.67 | 0.91 | 0.94 | −0.09 | 0.14 |
| tr|Q2FYY6|Q2FYY6_STAA8 - Glutamine synthetase OS=Staphylococcus aureus (strain NCTC 8325) GN=SAOUHSC_01287 PE=3 SV=1 xxxxx | 50841 | 6 | 9 | 2 | 10 | 4 | 2 | 5.67 | 5.33 | 0.92 | 0.94 | −0.09 | 0.12 |
Supplementary Material
Acknowledgments
C.S.L. was supported through a postdoctoral research fellowship by the German Research Foundation (DFG). This work was further supported through NIH grants 1R01GM111703 to M.B., 1R01GM117004 and 1R01GM118431-01A1 to E.W., 1R01AI101171 and 1R01AI069233 to E.P.S., and R21AI117255 to M.R.A. We thank Alexander Horswill (University of Iowa) for sharing the GFP plasmid pCM29. We thank Neri Amara and Josh Yim for help with NMR analyses and Shiyu Chen for LC-MS analysis of JCP678, and Lauren Popov, Oliwia Zurek, Joe Romaniuk and Lynette Cegelski for helpful discussions.
Footnotes
Author Contributions
C.S.L. and M.B. conceived the project. C.S.L designed and performed the in vitro experiments, synthesized compounds and analyzed data. J.R.S. designed and performed the in vivo infection experiments and the genetic manipulation of S. aureus, and analyzed data. L.A.C. and E.W. performed LC-MS/MS analysis. R.C. contributed to the comparative bacterial labeling experiments. M.G. synthesized compounds. M.A. contributed to the experimental design and analyzed data. E.P.S. designed and analyzed in vivo infection experiments. M.B. supervised the project, designed experiments and analyzed data. C.S.L. and M.B. wrote the manuscript, all authors reviewed, discussed and edited the manuscript.
Competing Interests: The authors declare that they have no competing interests as defined by Nature Research, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Data availability and Code availability statements
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009210 (https://www.ebi.ac.uk/pride/archive/).
References
- 1.Reddy PN, Srirama K, Dirisala VR. An Update on Clinical Burden, Diagnostic Tools, and Therapeutic Options of Staphylococcus aureus. Infect Dis (Auckl) 2017;10:1179916117703999. doi: 10.1177/1179916117703999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG., Jr Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–61. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laupland KB, et al. The changing epidemiology of Staphylococcus aureus bloodstream infection: a multinational population-based surveillance study. Clin Microbiol Infect. 2013;19:465–71. doi: 10.1111/j.1469-0691.2012.03903.x. [DOI] [PubMed] [Google Scholar]
- 4.Duthie ES, Lorenz LL. Staphylococcal coagulase; mode of action and antigenicity. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 5.Frees D, Gerth U, Ingmer H. Clp chaperones and proteases are central in stress survival, virulence and antibiotic resistance of Staphylococcus aureus. Int J Med Microbiol. 2014;304:142–9. doi: 10.1016/j.ijmm.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Staub I, Sieber SA. Beta-lactam probes as selective chemical-proteomic tools for the identification and functional characterization of resistance associated enzymes in MRSA. J Am Chem Soc. 2009;131:6271–6. doi: 10.1021/ja901304n. [DOI] [PubMed] [Google Scholar]
- 7.Frankel MB, Hendrickx AP, Missiakas DM, Schneewind O. LytN, a murein hydrolase in the cross-wall compartment of Staphylococcus aureus, is involved in proper bacterial growth and envelope assembly. J Biol Chem. 2011;286:32593–605. doi: 10.1074/jbc.M111.258863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukowski M, Wladyka B, Dubin G. Exfoliative toxins of Staphylococcus aureus. Toxins (Basel) 2010;2:1148–65. doi: 10.3390/toxins2051148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pietrocola G, Nobile G, Rindi S, Speziale P. Staphylococcus aureus Manipulates Innate Immunity through Own and Host-Expressed Proteases. Front Cell Infect Microbiol. 2017;7:166. doi: 10.3389/fcimb.2017.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottcher T, Sieber SA. Beta-lactones as specific inhibitors of ClpP attenuate the production of extracellular virulence factors of Staphylococcus aureus. J Am Chem Soc. 2008;130:14400–1. doi: 10.1021/ja8051365. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96:14694–9. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics. 2001;1:1067–71. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 13.Simon GM, Cravatt BF. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J Biol Chem. 2010;285:11051–5. doi: 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henness S, Perry CM. Orlistat: a review of its use in the management of obesity. Drugs. 2006;66:1625–56. doi: 10.2165/00003495-200666120-00012. [DOI] [PubMed] [Google Scholar]
- 15.Thornberry NA, Weber AE. Discovery of JANUVIA (Sitagliptin), a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. Curr Top Med Chem. 2007;7:557–68. doi: 10.2174/156802607780091028. [DOI] [PubMed] [Google Scholar]
- 16.Kluge AF, Petter RC. Acylating drugs: redesigning natural covalent inhibitors. Curr Opin Chem Biol. 2010;14:421–7. doi: 10.1016/j.cbpa.2010.03.035. [DOI] [PubMed] [Google Scholar]
- 17.Adam GC, Sorensen EJ, Cravatt BF. Chemical strategies for functional proteomics. Mol Cell Proteomics. 2002;1:781–90. doi: 10.1074/mcp.r200006-mcp200. [DOI] [PubMed] [Google Scholar]
- 18.Leung D, Hardouin C, Boger DL, Cravatt BF. Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat Biotechnol. 2003;21:687–91. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- 19.Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Acad Sci U S A. 2002;99:10335–40. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ortega C, et al. Systematic Survey of Serine Hydrolase Activity in Mycobacterium tuberculosis Defines Changes Associated with Persistence. Cell Chem Biol. 2016;23:290–298. doi: 10.1016/j.chembiol.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tallman KR, Levine SR, Beatty KE. Small-Molecule Probes Reveal Esterases with Persistent Activity in Dormant and Reactivating Mycobacterium tuberculosis. ACS Infect Dis. 2016;2:936–944. doi: 10.1021/acsinfecdis.6b00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatzios SK, et al. Chemoproteomic profiling of host and pathogen enzymes active in cholera. Nat Chem Biol. 2016;12:268–274. doi: 10.1038/nchembio.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zweerink S, et al. Activity-based protein profiling as a robust method for enzyme identification and screening in extremophilic Archaea. Nat Commun. 2017;8:15352. doi: 10.1038/ncomms15352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall CI, et al. Chemical genetic screen identifies Toxoplasma DJ-1 as a regulator of parasite secretion, attachment, and invasion. Proc Natl Acad Sci U S A. 2011;108:10568–73. doi: 10.1073/pnas.1105622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lentz CS, et al. Design of Selective Substrates and Activity-Based Probes for Hydrolase Important for Pathogenesis 1 (HIP1) from Mycobacterium tuberculosis. ACS Infect Dis. 2016;2:807–815. doi: 10.1021/acsinfecdis.6b00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cadieux B, Vijayakumaran V, Bernards MA, McGavin MJ, Heinrichs DE. Role of lipase from community-associated methicillin-resistant Staphylococcus aureus strain USA300 in hydrolyzing triglycerides into growth-inhibitory free fatty acids. J Bacteriol. 2014;196:4044–56. doi: 10.1128/JB.02044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenstein R, Gotz F. Staphylococcal lipases: biochemical and molecular characterization. Biochimie. 2000;82:1005–14. doi: 10.1016/s0300-9084(00)01180-9. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen MT, et al. Staphylococcal (phospho)lipases promote biofilm formation and host cell invasion. Int J Med Microbiol. 2017 doi: 10.1016/j.ijmm.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Staub I, Sieber SA. Beta-lactams as selective chemical probes for the in vivo labeling of bacterial enzymes involved in cell wall biosynthesis, antibiotic resistance, and virulence. J Am Chem Soc. 2008;130:13400–9. doi: 10.1021/ja803349j. [DOI] [PubMed] [Google Scholar]
- 30.Fey PD, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4:e00537–12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Utaida S, et al. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology. 2003;149:2719–32. doi: 10.1099/mic.0.26426-0. [DOI] [PubMed] [Google Scholar]
- 32.Pietiainen M, et al. Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics. 2009;10:429. doi: 10.1186/1471-2164-10-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sass P, et al. The lantibiotic mersacidin is a strong inducer of the cell wall stress response of Staphylococcus aureus. BMC Microbiol. 2008;8:186. doi: 10.1186/1471-2180-8-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muthaiyan A, et al. Antimicrobial effect and mode of action of terpeneless cold-pressed Valencia orange essential oil on methicillin-resistant Staphylococcus aureus. J Appl Microbiol. 2012;112:1020–33. doi: 10.1111/j.1365-2672.2012.05270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palazzolo-Ballance AM, et al. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J Immunol. 2008;180:500–9. doi: 10.4049/jimmunol.180.1.500. [DOI] [PubMed] [Google Scholar]
- 36.Bore E, Langsrud S, Langsrud O, Rode TM, Holck A. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology. 2007;153:2289–303. doi: 10.1099/mic.0.2007/005942-0. [DOI] [PubMed] [Google Scholar]
- 37.Neumann Y, et al. The effect of skin fatty acids on Staphylococcus aureus. Arch Microbiol. 2015;197:245–67. doi: 10.1007/s00203-014-1048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garland M, et al. Development of an activity-based probe for acyl-protein thioesterases. PLoS One. 2018;13:e0190255. doi: 10.1371/journal.pone.0190255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jinno A, Park PW. Role of glycosaminoglycans in infectious disease. Methods Mol Biol. 2015;1229:567–85. doi: 10.1007/978-1-4939-1714-3_45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Gemst JJ, et al. RNA Contaminates Glycosaminoglycans Extracted from Cells and Tissues. PLoS One. 2016;11:e0167336. doi: 10.1371/journal.pone.0167336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raz A, Fischetti VA. Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc Natl Acad Sci U S A. 2008;105:18549–54. doi: 10.1073/pnas.0808301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci U S A. 2000;97:5510–5. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clarke AJ, Dupont C. O-acetylated peptidoglycan: its occurrence, pathobiological significance, and biosynthesis. Can J Microbiol. 1992;38:85–91. doi: 10.1139/m92-014. [DOI] [PubMed] [Google Scholar]
- 44.Bera A, Biswas R, Herbert S, Gotz F. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect Immun. 2006;74:4598–604. doi: 10.1128/IAI.00301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown S, et al. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc Natl Acad Sci U S A. 2012;109:18909–14. doi: 10.1073/pnas.1209126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winstel V, et al. Wall Teichoic Acid Glycosylation Governs Staphylococcus aureus Nasal Colonization. MBio. 2015;6:e00632. doi: 10.1128/mBio.00632-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weadge JT, Clarke AJ. Identification and characterization of O-acetylpeptidoglycan esterase: a novel enzyme discovered in Neisseria gonorrhoeae. Biochemistry. 2006;45:839–51. doi: 10.1021/bi051679s. [DOI] [PubMed] [Google Scholar]
- 48.Weadge JT, Pfeffer JM, Clarke AJ. Identification of a new family of enzymes with potential O-acetylpeptidoglycan esterase activity in both Gram-positive and Gram-negative bacteria. BMC Microbiol. 2005;5:49. doi: 10.1186/1471-2180-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rahman MM, et al. The Staphylococcus aureus Methicillin Resistance Factor FmtA Is a d-Amino Esterase That Acts on Teichoic Acids. MBio. 2016;7:e02070–15. doi: 10.1128/mBio.02070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia-Fernandez E, et al. Membrane Microdomain Disassembly Inhibits MRSA Antibiotic Resistance. Cell. 2017;171:1354–1367e20. doi: 10.1016/j.cell.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pang YY, et al. agr-Dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J Innate Immun. 2010;2:546–559. doi: 10.1159/000319855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.