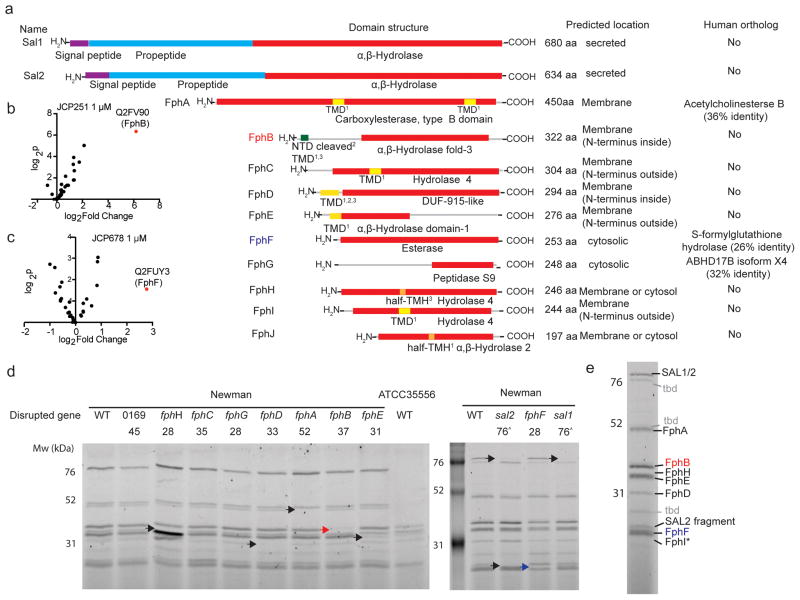
Fig. 2. LC-MS/MS-based identification of serine hydrolases in S. aureus.
a) Domain structure prediction and bioinformatics analyses of identified hydrolases. Red: α,β-Hydrolase domain (family specification according to Pfam). Purple: signal peptides. Cyan: propeptides. Yellow: transmembrane domain (TMD). Orange: Possible half-transmembrane-helix (half-TMH). Green: Ambiguous N-terminal domain (NTD). The programs predicting the presence of a TMD/TMH are indicated as follows: 1 TmPred, 2Phobius, 3MemBrain. For identification of human orthologs by blastp, the non-redundant protein sequences database for Homo sapiens was queried using the indicated full-length protein sequences. b) Volcano plot of FP-biotin targets in S. aureus ATCC35556 showing change in recovery upon JCP251 and c) JCP678 pretreatment relative to a control. The top hit (highest p value and most significant change in recovery) is labeled. P-values were calculated using a two-tailed t-test on n=3 biologically independent samples for each of the two groups being compared. d) FP-TMR labeling profiles of S. aureus Newman transposon mutant strains with insertions in serine hydrolase genes. Arrowheads indicate labeled proteins disappearing in individual mutant strains (Red arrowhead: FphB, blue arrowhead: FphH). The experiment was performed twice with similar results. e) The primary labeled targets of FP-TMR in S. aureus Newman. Identities of species confirmed by mutational analysis are indicated, * FphI identity is predicted based on molecular weight. ‘tbd’ identity remains unconfirmed