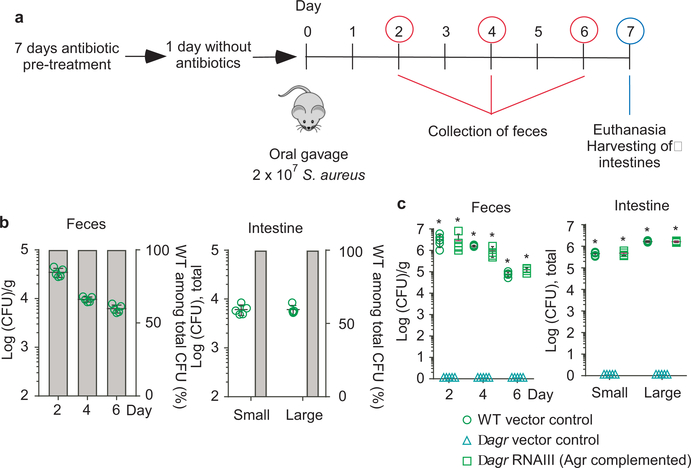
Figure 2 |. Quorum-sensing dependence of S. aureus intestinal colonization.
a, Experimental setup of the murine intestinal colonization model. Mice received by oral gavage 100 μl containing 108 CFU/ml of wild-type (WT) S. aureus strain ST2196 F12 and another 100 μl of 108 CFU/ml of the corresponding isogenic agr mutant (n=5/group, competitive experiment shown in b), or 200 μl containing 108 CFU/ml wild-type, isogenic agr mutant, or Agr (RNAIII)-complemented agr mutant (n=5/group, non-competitive experiment shown in c). CFU in the feces were determined two, four, and six days after infection. At the end of the experiment (day seven), CFU in the small and large intestines were determined. b, Competitive experiment. Total obtained CFU are shown as dot plots, also showing the means ±SD. Bars show the percentage of wild-type among total determined CFU, of which 100 were analyzed for tetracycline resistance that is present only in the agr mutant. No agr mutants were detected in any experiment; thus, all bars show 100%. Given that 100 isolates were tested, the competitive index wild-type/agr mutant in all cases is ≥ 100. c, Non-competitive experiment with genetically complemented strains. Wild-type and isogenic agr mutant strains all harbored the pKXΔ16 control plasmid; Agr-complemented strains harbored pKXΔRNAIII, constitutively expressing RNAIII, which is the intracellular effector of Agr. During the experiment, mice received 200 μg/ml kanamycin in the drinking water to maintain plasmids. Statistical analysis was performed using Poisson regression versus values obtained with the agr mutant strains. *, p<0.0001. Error bars shown the means ±SD. Note no bacteria were found in the feces or intestines of any mouse receiving S. aureus Δagr with vector control. The corresponding zero values are plotted on the x-axis of the logarithmic scale. See Extended Data Fig. 2 for the corresponding data using strains USA300 LAC and ST88 JSNZ.