Figure 5. In silico docking to design degrader molecules.
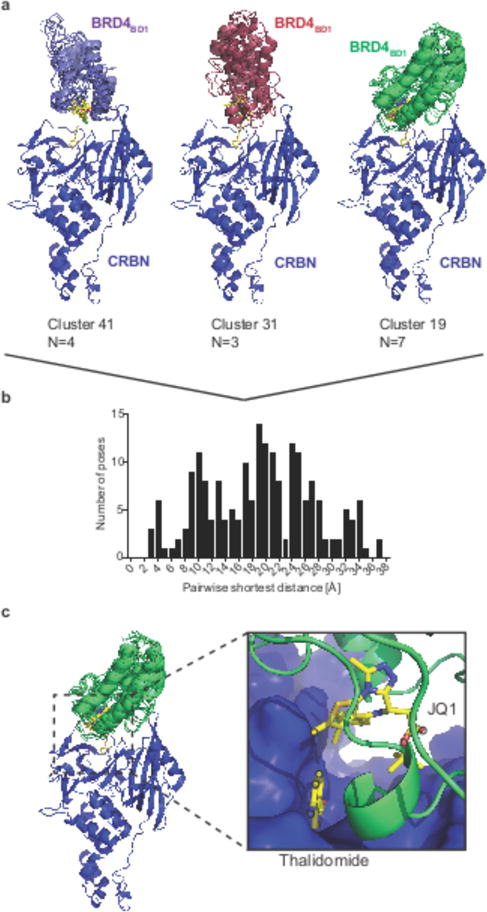
(a) Cartoon representations for representative clusters obtained by k-means clustering of the top 200 global docking poses between CRBN (pdb: 4tz4) and BRD4BD1 (pdb: 3mxf). (b) Histogram of the pairwise shortest distances for the top 200 docking poses. (c) Close-up view on the proximity of the JQ1 thiophene and lenalidomide that provided the rationale for synthesizing ZXH-2-147 and ZXH-3-26. Atoms used for calculation of the pairwise shortest distances between JQ1 and lenalidomide are highlighted in black circles.
