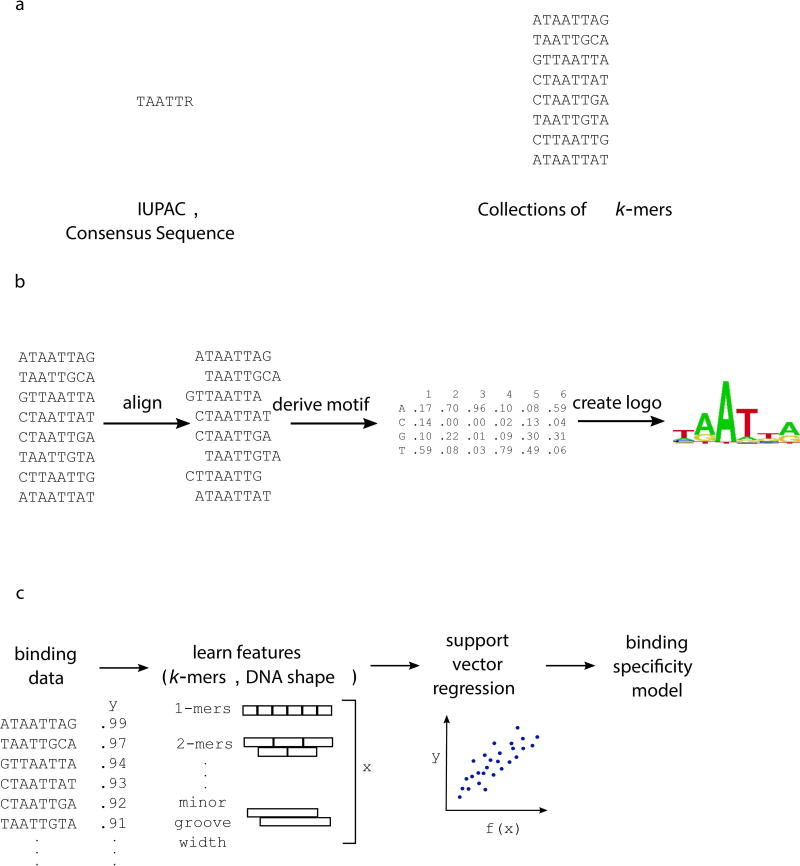
Figure 1. Representations of binding specificity.
(a) Groups of sequences bound by a TF can be used to create a consensus sequence, represented using IUPAC notation. The group of k-mers themselves can be used to denote sequences bound by the TF. (b) Here, bound sequences are aligned to create a motif, which indicates the probability of each nucleotide at every position within the binding site. Multiple algorithms exist for creating a PWM from high-throughput binding data (reviewed in Stormo, 2013). (c) Machine learning approaches can learn specificity models from binding data, incorporating short k-mer and DNA shape features of the DNA binding sites.