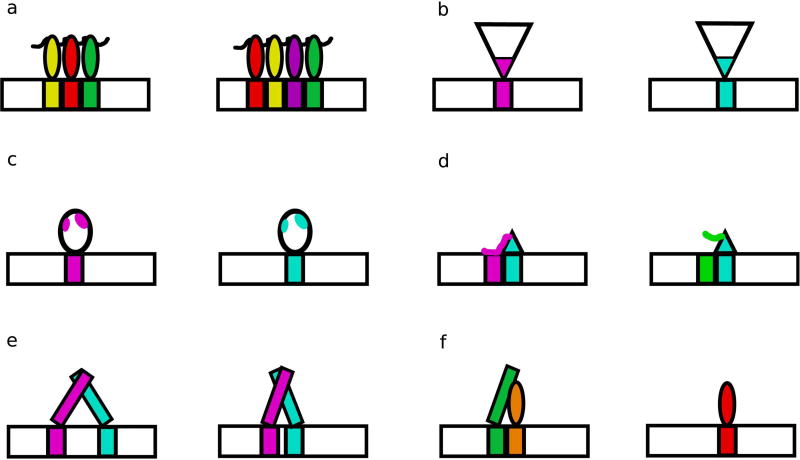
Figure 2. Structural differences between TFs enable divergence in DNA binding.
(a) Modular TF families, such as ZFs, contain members with different numbers and arrangements of individual DBDs. (b) Members of a family can contain different amino acids at DNA-contacting positions, as seen in both the fly and mouse HD specificity classes (Berger et al., 2008; Noyes et al., 2008). (c) Differences not at DNA-contacting positions can alter specificity through allosteric mechanisms, as observed in the human ETS factors SAP-1 and Elk-1 (Mo et al., 2000). (d) Protein loops can contact DNA flanking the core recognition motif, adding preferences for DNA shape features, as seen in the yeast S. cerevisiae bHLH proteins Cbf1 and Tye7 (Gordan et al., 2013). (e) DBDs that bind as dimers can recognize sites with different spacer lengths between half sites, as seen in yeast S. cerevisiae bZIP proteins (Gordan et al., 2011). (f) DNA binding along with a co-factor can change the specificity of a TF, as observed in the specificities of the fly Hox protein binding with the cofactors Exd and Hth (Slattery et al., 2011).