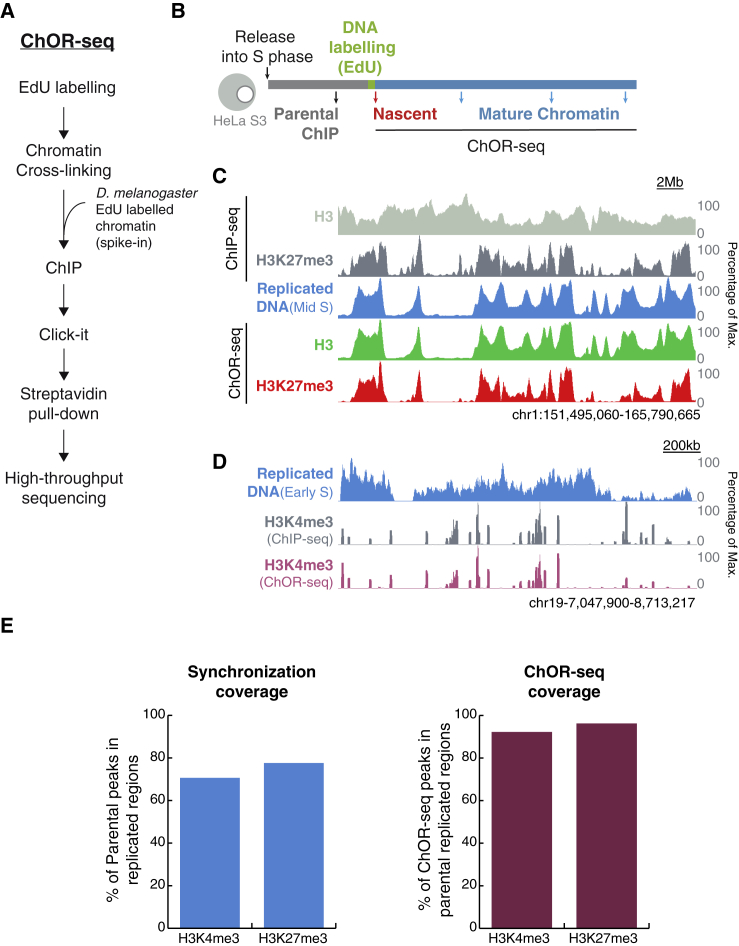
Figure 1.
Tracking Histone PTM Occupancy after DNA Replication with ChOR-Seq
(A) Overview of the ChOR-seq protocol.
(B) Experimental setup. HeLa S3 cells were released into S phase from a thymidine block. Parental and nascent chromatin were collected 1 hr before or immediately after EdU labeling, respectively. The EdU label was then chased and mature chromatin harvested at selected time points along the cell cycle.
(C and D) Parental ChIP-seq and nascent ChOR-seq profiles of pan-H3 and H3K27me3 (C) and H3K4me3 (D). Replicated DNA profiles are shown in blue. Signal is scaled as percentage of maximum at the locus depicted.
(E) Bar plots showing the synchronization coverage (left) and ChOR-seq coverage (right) in the H3K4me3 and H3K27me3 datasets. Percentage is calculated from peaks subsetted into 500 bp non-overlapping windows.
See also Figure S1.