Figure 2.
The Histone H3 PTM Landscape Is Accurately Reproduced upon Replication of Active and Repressed Genomic Loci
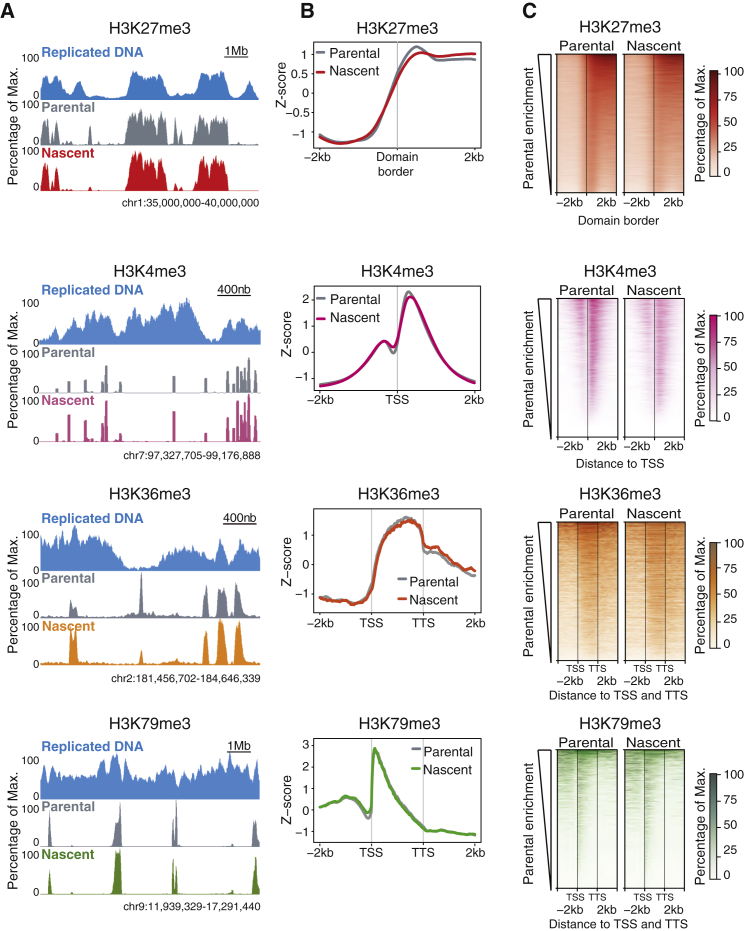
(A) Histone PTM profiles from ChIP-seq (parental) and ChOR-seq (nascent) of H3K27me3, H3K4me3, H3K36me3, and H3K79me3. Replicated DNA profiles are shown in blue. Signal is scaled as percentage of maximum at the locus depicted.
(B) Average profiles of parental and nascent H3K27me3, H3K4me3, H3K36me3, and H3K79me3. H3K27me3 signal is plotted across 4 kb centered on borders of replicated H3K27me3 domains. H3K4me3 signal is plotted across 4 kb centered on replicated TSSs. H3K36me3 and H3K79me3 signal is plotted from 2 kb upstream to 2 kb downstream of replicated open reading frames. All data shown is Z score normalized.
(C) Heatmaps of parental and nascent H3K27me3, H3K4me3, H3K36me3, and H3K79me3 signal across the regions described in (B). Color intensity represents percentage of maximum levels set separately for parental and nascent samples.
See also Figure S2.