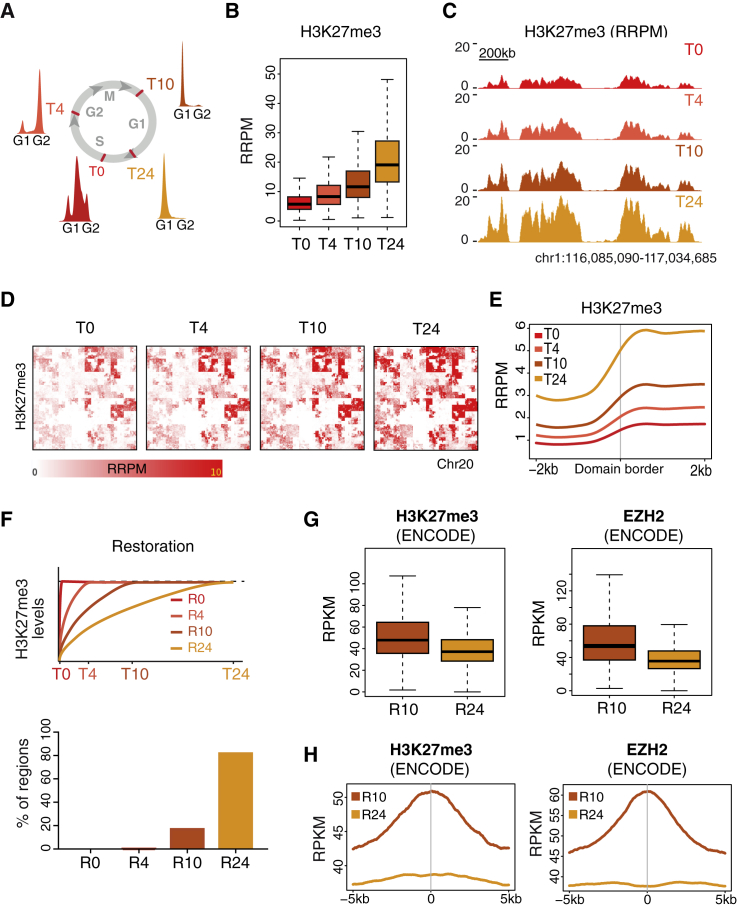
Figure 4.
High PRC2 Occupancy Sites Show Faster H3K27me3 Restoration
(A) Outline of H3K27me3 qChOR-seq time course analysis. Cell cycle progression was monitored by FACS analysis of DNA content.
(B) Boxplots of H3K27me3 qChOR-seq signal in replicated parental peaks subsetted into 2 kb non-overlapping windows.
(C) Comparison of H3K27me3 nascent and mature qChOR-seq profiles.
(D) Hilbert curves of H3K27me3 qChOR-seq signal over chromosome 20 at the indicated time points. Colored areas reflect the size and signal of H3K27me3-enriched domains.
(E) Average profiles of H3K27me3 qChOR-seq signal across 4 kb centered on the border of replicated H3K27me3 domains.
In (B)–(E), signal is quantitated using reference-adjusted reads per million (RRPM).
(F) Top: scheme of strategy used to parse H3K27me3-enriched regions by restoration kinetics. Regions were defined as R0, R4, R10, or R24 based on the time point at which R24 H3K27me3 levels were reached. Bottom: bar chart of the proportion of H3K27me3-enriched regions in each restoration category. Regions are defined as 2 kb non-overlapping windows in replicated parental peaks.
(G) Boxplots of ENCODE H3K27me3 signal (left) and ENCODE EZH2 signal (right) in R10 and R24 regions. Signal is quantitated using reads per kilobase per million (RPKM).
(H) Average profiles of ENCODE H3K27me3 signal (left) and ENCODE EZH2 signal (right) across 10 kb centered on R10 and R24 regions.
See also Figure S4.