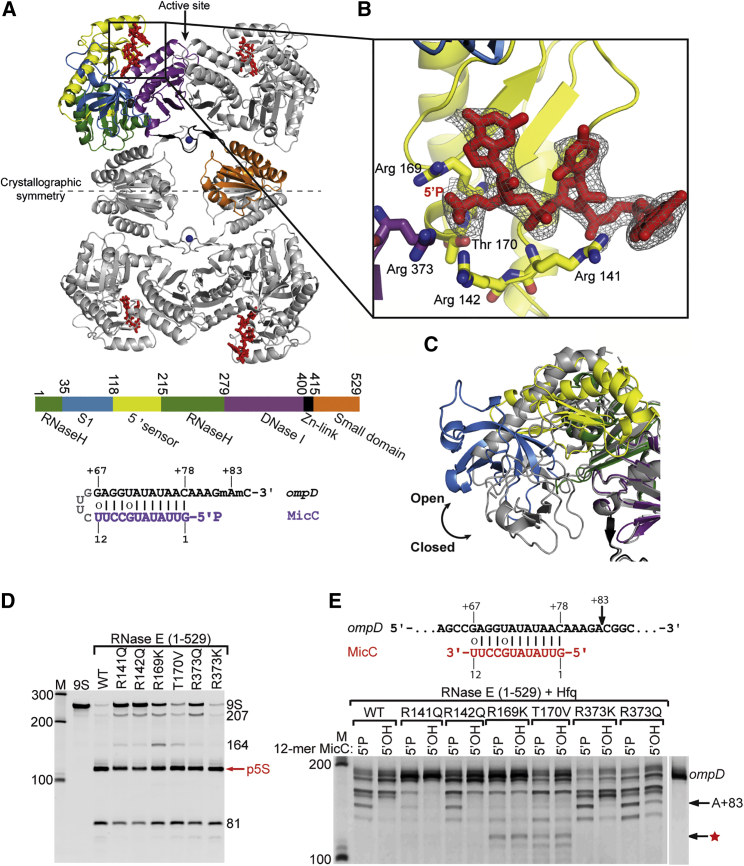
Figure 1.
The Structure of RNase E with a Fragment of MicC sRNA Identifies Residues Involved in 5′ End Recognition
(A) RNase E (1–529)-MicC quaternary structure. The individual subdomains of one protomer are color coded as indicated in the coding bar. Bound RNA fragment (red), interacting with 5′ sensor and S1 domains, is colored on each subunit. The schematic of RNA substrate used for crystallization is shown in the bottom of the panel. The lines indicate predicted pairings and the circles wobble-pairs.
(B) Contacts to the 5′ end of the RNA. The ternary and quaternary structural changes as well as the 5′ end sensor domain contacts to the RNA are corroborated by another structure of RNase E at 3.5–3.7 Å resolution crystallized in a different space group with 1.5 tetramers in the asymmetric unit (data not shown). The anneal-omit map (Fo-Fc coefficients) was calculated using the final coordinates and contoured at 2.3 sigma.
(C) Tertiary structural changes associated with closed (2c4R, gray) and open (color coded as in A) states for a protomer.
(D) The role of the amino acids in- and outside of the 5′ binding pocket in processing 9S rRNA. The red arrow indicates 5S precursor (p5S), the final product of 9S processing.
(E) The role of the amino acids in- and outside of the 5′ binding pocket in MicC-mediated ompD degradation. RNase E (1–529) wild-type (WT) and mutants (R141Q, R142Q, R169K, T170V, R373Q, and R373K) were evaluated with 200 nM 9S or 200 nM ompD in the presence of 300 nM 12-mer MicC. For each enzyme, the concentration used was 200 nM for 9S assays and 150 nM for MicC/ompD assays. The size markers are in the left lanes (M). The black arrow indicates the 153 nt long product of the +83 cleavage of ompD, which is the in vivo observed MicC-guided cleavage. The star indicates a new cleavage site observed for RNase E (1–529) R169K and T170V.