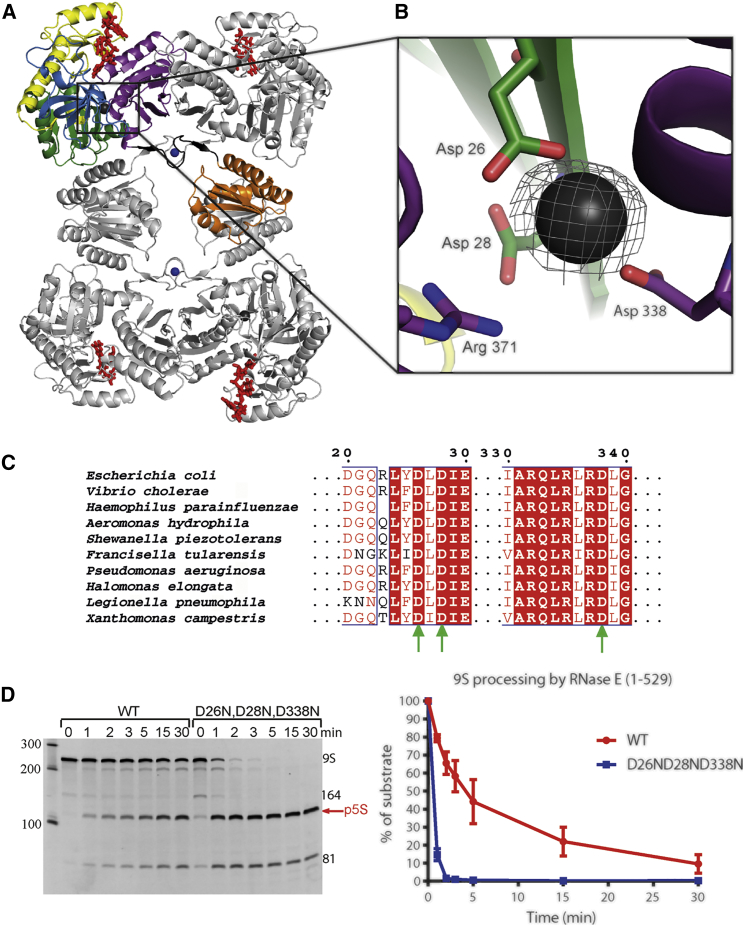
Figure 2.
Acidic Pocket in RNase E NTD Coordinates Magnesium Ion and Affects Activity of the Enzyme
(A and B) The conserved acidic region in RNase E (A) and a putative Mg(II) interaction (B). One protomer of RNase E is color coded as in Figure 1. The Fo-Fc anneal-omit map for the magnesium ion was calculated using the phases from the final coordinates and contoured at 2.5 sigma.
(C) Alignment of RNase E catalytic domain from representative species of γ-Proteobacteria. Green arrows mark residues coordinating the metal in the newly identified magnesium binding site.
(D) Substitution of the conserved aspartates with asparagines (RNase E D26N,D28N,D338N) tremendously boosts the activity of the enzyme for processing 9S RNA. In total, 200 nM 9S was incubated with 50 nM RNase E (1–529) wild-type (WT) or the D26N,D28N,D338N mutant. The profiles show averages and SDs from three technical replicates.