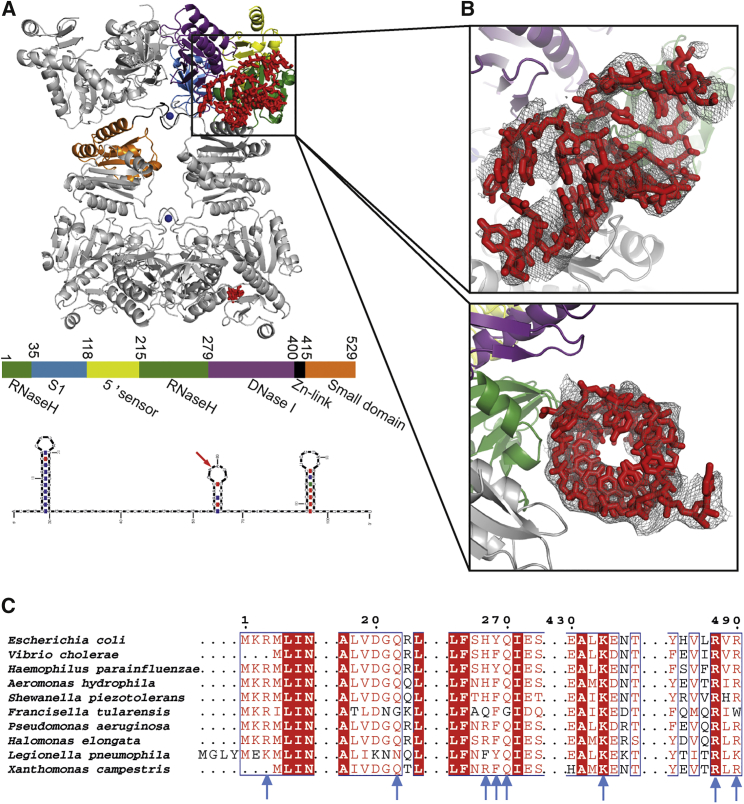
Figure 4.
Crystal Structure of the Complex of sRNA RprA with RNase E Catalytic Domain
(A) Refined structure of RNase E (1–511) showing the duplex binding on the surface of the RNase H domain and the small domain of the partner protomer of the principal dimer. The helix is proposed to be from the 5′ stem-loop structure of RprA (bottom panel). The individual subdomains of one protomer are color coded as indicated in the coding bar. RNA is shown in red.
(B) RNA interacting with the duplex binding surface. Fo-Fc anneal-omit electron density map showing putative duplex region and with fitted RNA stem-loop structure was calculated using the final coordinates and contoured at 1.5 sigma.
(C) Alignment of RNase E catalytic domain from representative species of γ-Proteobacteria. Blue arrows mark residues implicated in interaction with RNA structural elements based on the crystal structure of the RNase E NTD/RprA complex.