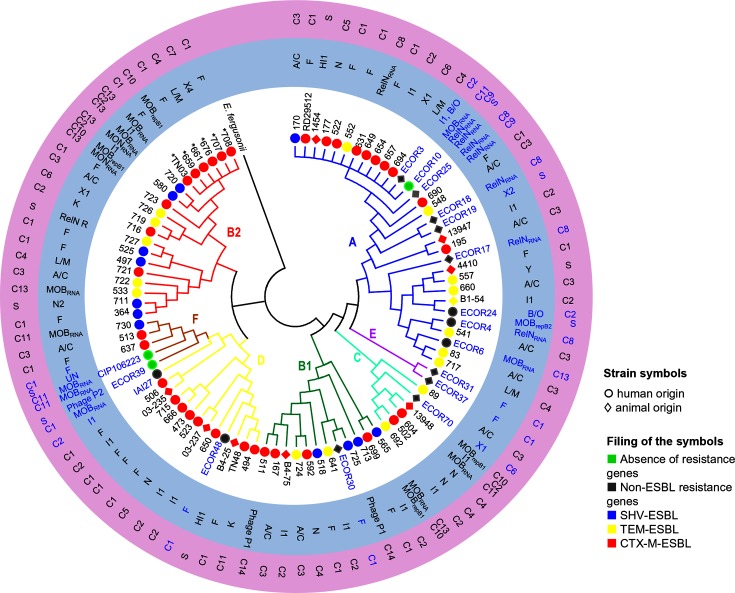
Fig. 5.
Phylogeny of the E. coli host strains. Phylogenetic tree reconstructed from the alignment of the concatenated sequences of eight genes of the MLST IP scheme using PhyML under the GTR model, with E. fergusonii as the outgroup. The phylogroups are indicated with the colours blue (A), green (B1), red (B2), turquoise (C), yellow (D), purple (E) and brown (F). First circle (no background): strain ID in blue for the strains isolated before the use of 3GCs and in black for the strains isolated after the use of 3GCs. * indicates strains of ST43 (ST131 Achtman MLST scheme); symbols indicate strain origin, fill colour of the symbols indicates the type of resistance (red, blue and yellow are for strains carrying at least one ESBL-encoding gene). Second circle (blue background): corresponding plasmids studied for each strain, MPF plasmids are indicated by their Inc group, and MOB plasmids and RelN plasmids by the type of replication system. Third circle (purple background): cluster number (C1 to C13) to which the plasmids belong, as defined in Fig. 4, and S (singleton) for plasmids not linked to a cluster.