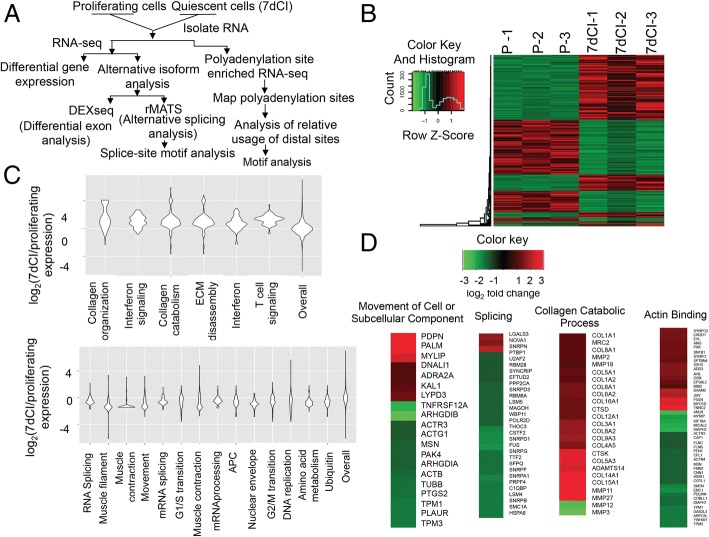
Fig. 1.
RNA-Seq analysis of gene expression changes in proliferating versus quiescent fibroblasts. a Schematic of RNA-Seq-based analysis of proliferating and quiescent fibroblasts performed in this study. b Total RNA was isolated from three independent biological replicates of proliferating fibroblasts and three matched independent biological replicates of 7dCI fibroblasts. RNA samples were converted to cDNA libraries and sequenced on an Illumina Hi-Seq 2000. Reads were aligned to the human genome (hg19 human reference sequence) and the number of reads mapping to each gene (UCSC gene annotation) in the genome was determined. A heatmap of read counts for 1993 genes with at least a twofold change in expression and a false discovery rate (FDR) < 5% is shown. Hierarchical clustering is denoted by the dendrogram to the left of the heatmap. A color key and a histogram displaying the density of genes at a given color intensity are shown in the upper left corner. c Gene set enrichment analysis was used to determine the gene sets most significantly upregulated (top) or downregulated (bottom) with quiescence. Gene sets are listed in descending order of statistical significance from left to right. A histogram of the log2(fold-change) of the normalized read count in 7dCI compared to proliferating fibroblasts for each gene in the gene set is plotted in a violin plot representation. d Heat maps of genes within selected gene set enrichment categories are provided. The log2 ratio of normalized RNA-Seq counts in 7dCI compared with proliferating fibroblasts are shown. Red indicates higher expression in quiescent than proliferating fibroblasts; green indicates higher expression in proliferating than quiescent fibroblasts. Only genes in each category that change in expression two-fold or more are included