Figure 3. IpRGCs express ion channels regulated by cyclic nucleotide and produce cyclic nucleotide in phototransduction.
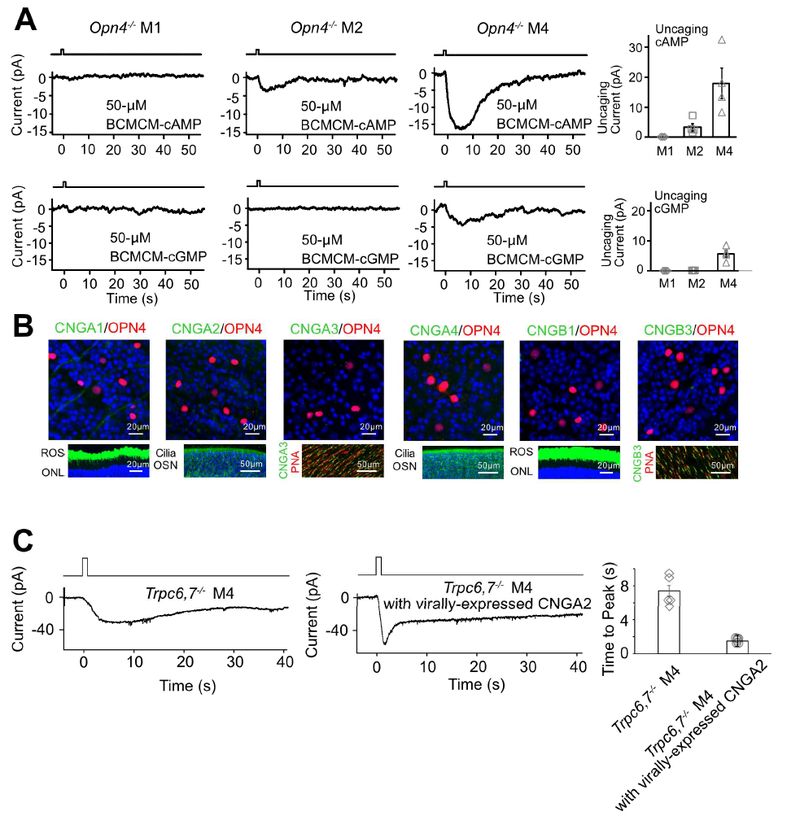
(A) Opn4−/− genotype. Voltage-clamp recordings at −66 mV. Left 6 panels, Inward currents elicited by photo-uncaged cAMP or cGMP in M1-, M2- and M4-cells. 50-μM BCMCM-cAMP or -cGMP was loaded into the recorded cell via the whole-cell pipette. Photo-uncaging achieved by white-light spot (Hg lamp, 1 sec, 0.12 μW μm−2, 40μm in diameter) centered on recorded soma. Far right, Collected data (mean ± SEM, n =3, 4, 4 cells at top and 4, 4, 4 cells at bottom from at least 2 animals in each group; see text). (B) Top, Lack of immunosignals for various CNGA- and CNGB-subunits (Methods) in RGC layer of flat-mount Opn4-Cre;Rosa-tdTomato retina (red, tdTomato indicating ipRGCs; green, respective CNG channel but not detected). Bottom, Verification of positive immunostaining (green) with the same CNG-antibodies for rods and cones as well as olfactory sensory neurons. Retinal cross-sections for CNGA1 and CNGB1; ROS, rod outer segment; ONL, outer nuclear layer. Main olfactory epithelial cross-sections for CNGA2 and CNGA4; OSN, olfactory sensory neurons with cilia. Flat-mount retinas for CNGA3 and CNGB3; cone outer segments marked by Peanut Agglutinin (PNA, red). Blue color is DAPI nuclear staining. (C) Trpc6,7−/− M4-ipRGC with virally-expressed CNGA2 subunit (middle) has a light response with a faster rising phase compared to non-infected cell (left), indicating that cyclic nucleotide is produced in the unknown phototransduction pathway. The duration of light stimulation was 1 second. Right, Collected data of time to peak, mean ± SEM, n = 5, 6 cells from at least 2 animals in each group. See also Figure S3.
