Abstract
Myelin transcription factor 1 (Myt1) and Myt1l (Myt1-like) are zinc finger transcription factors that regulate neuronal differentiation. Reduced Myt1l expression has been implicated in glioblastoma (GBM), and the related St18 was originally identified as a potential tumor suppressor for breast cancer. We previously analyzed changes in gene expression in a human GBM cell line with re-expression of either Myt1 or Myt1l. This revealed largely overlapping gene expression changes, suggesting similar function in these cells. Here we show that re-expression of Myt1 or Myt1l reduces proliferation in two different GBM cell lines, activates gene expression programs associated with neuronal differentiation, and limits expression of proliferative and epithelial to mesenchymal transition gene-sets. Consistent with this, expression of both MYT1 and MYT1L is lower in more aggressive glioma sub-types. Examination of the gene expression changes in cells expressing Myt1 or Myt1l suggests that both repress expression of the YAP1 transcriptional coactivator, which functions primarily in the Hippo signaling pathway. Expression of YAP1 and its target genes is reduced in Mytexpressing cells, and there is an inverse correlation between YAP1 and MYT1/MYT1L expression in human brain cancer datasets. Proliferation of GBM cell lines is reduced by lowering YAP1 expression and increased with YAP1 over-expression, which overcomes the anti-proliferative effect of Myt1/Myt1l expression. Finally we show that reducing YAP1 expression in a GBM cell line slows the growth of orthotopic tumor xenografts. Together, our data suggest that Myt1 and Myt1l directly repress expression of YAP1, a protein which promotes proliferation and GBM growth.
Keywords: Gene expression, Transcription, Repression, Cancer, Glioma, Proliferation
Glioblastoma (GBM) is a highly aggressive cancer with poor prognosis. GBM is the most frequent primary malignant brain cancer in adults, yet due to intra-tumor heterogeneity and its resistance to common therapies, GBM treatment options are relatively limited [1, 2]. Median survival for GBM patients remains below two years, even if patients initially respond to standard therapies, including surgical resection, radiation and chemotherapies, such as temozolomide treatment [3]. Recent molecular profiling of GBM has identified common alterations in known oncogenes and tumor suppressors, including PTEN, EGFR, and TP53, among others [4, 5]. In addition, these analyses classified GBMs into various subtypes that may be more clinically relevant than previous histological grades. However, despite advances in understanding the underlying nature of GBM and its inherent heterogeneity, effective treatment options are still quite limited.
Myt1 and Myt1l (Myelin transcription factor 1 and Myt1-like) are two of the three members of a small family of zinc finger transcription factors that are characterized by two clusters of C2HC zinc fingers [6, 7]. The DNA sequence to which these fingers bind has been defined in vitro [8, 9], and evidence from genome wide analyses supports that this is responsible for at least part of the targeting of these factors to DNA [10, 11]. Characterization of the transcriptional activity of the Myt protein family has suggested that they play roles in repression and activation. Myt1 represses transcription, interacts with Sin3 and histone deacetylases, and can be incorporated into a neural specific LSD1 containing corepressor complex [12, 13]. In contrast, Myt1l and a Xenopus homolog of Myt1 have been shown to activate transcription from synthetic reporters based on the known consensus site [6, 8]. However, recent evidence suggests that although there are potential differences in activity, regulation of direct target gene expression via the consensus site primarily results in transcriptional repression [11].
Myt family members are relatively widely expressed in neural tissue during embryonic development, but in the adult, expression is primarily limited to brain [14]. All three members of the Myt family have been implicated in neuronal differentiation in P19 cells, with transient overexpression promoting the conversion of undifferentiated P19 cells to neurons [15]. Myt1l, when expressed together with the transcription factors Brn2 and Ascl1, can reprogram fibroblasts to neurons [16–18]. In this context, Myt1l may function to limit non-neuronal gene expression programs and non-neuronal cell fate [10]. Thus expression of Myt family proteins in adult brain may function to maintain neural identify. St18 (Suppressor of tumorigenicity 18), which is the third member of this zinc finger family, was originally identified as a potential breast cancer tumor suppressor, suggesting that these proteins may play roles in tumorigenesis [19]. MYT1L has been shown to limit the growth of glioblastoma in a xenograft model, and was suggested to do so by regulating expression of the RNA binding protein, Rbfox1 [20]. In addition, there is evidence suggesting that higher MYT1L expression correlates with better patient survival in neuroblastoma, but otherwise little is known about potential functional roles of MYT1 and MYT1L in cancer [21].
The Hippo signaling pathway was first discovered through mutant screens in D. melanogaster, where mutations in several components resulted in similar overgrowth phenotypes [22–24]. Signaling via this pathway limits cellular proliferation and promotes cell death and differentiation. Although identified in flies, the core components of the pathway are all conserved in vertebrates, including humans [25]. Signaling via the Hippo pathway is regulated by inputs from planar cell polarity, apical-basal polarity and other stress cues, such as mechanical force. In mammalian cells, the MST1/2 kinases phosphorylate and activate a complex that includes LATS1/2 kinases, which then phosphorylate the YAP1/TAZ transcriptional co-activators [25]. YAP1 and TAZ (homologs of the fly Yorkie) are recruited to DNA indirectly via interaction with the TEAD DNA binding proteins (TEAD1–4, or Scalloped in Drosophila) [26]. While this represents the canonical Hippo pathway, YAP1/TAZ can also interact with other DNA-binding transcription factors, such as the SMAD proteins that mediate Transforming Growth Factor ß family signaling [27, 28]. Recruitment of YAP1/TAZ by TEAD results in activation of target gene expression, and this is inhibited by the upstream Hippo signaling pathway. Phosphorylation of YAP1/TAZ promotes interaction with 14–3-3 proteins and cytoplasmic sequestration [29–31], and can also lead to enhanced degradation of YAP1 and TAZ [32, 33]. Thus in the absence of signaling YAP1/TAZ drive pro-proliferative and anti-differentiation gene expression programs. Given the effects of Hippo signaling on cell survival and proliferation, deregulation of this pathway in cancer might be expected. The NF2 gene, which is an upstream activator of the Hippo pathway, has been identified as a tumor suppressor gene that is mutated in Type 2 neurofibromatosis, an autosomal dominant syndrome that results in meningioma and schwannoma [34, 35]. However, other pathway components appear not to be frequently mutated in cancer, although altered expression is seen, and hyperactivity of YAP1 and TAZ is not uncommon.
YAP1 expression is relatively high in proliferating cells in fetal brain, with much lower expression seen in adults [36]. In contrast, YAP1 expression is higher in GBM than in normal tissue, and inhibition of YAP1 activity has been suggested as a potential therapeutic approach in glioma [36–38]. In addition, TAZ activity has been implicated both in GBM tumorigenesis and the progression to high grade mesenchymal gliomas [39]. This role of YAP1/TAZ fits with the pro-proliferative and anti-differentiation functions of YAP1/TAZ in neural tissue in both mammals and flies [40–42].
Here we show that the zinc finger transcription factors, Myt1 and Myt1l reduce proliferation of human GBM cell lines and promote expression of gene sets associated with neural differentiation. YAP1 is among the genes repressed by expression of Myt1 and Myt1l, and consistent with this repression of YAP1 expression, Myt1/Myt1l and YAP1 have inverse expression patterns in GBM and lower grade glioma (LGG). Reducing YAP1 expression mimics the effect of Myt1/Myt1l re-expression, and YAP1 over-expression can block the anti-proliferative effects of Myt1 and Myt1l in GBM cell lines. Finally, we show that reducing YAP1 expression in GBM cells decreases tumor growth in a xenograft model, and we propose a model in which Myt1/Myt1l directly repress YAP1 expression to limit proliferation and tumor growth.
MATERIALS AND METHODS
Cell culture, transfection, lentiviral infection and western blot
HEK293T, A172, U87 and SKNSH cells were maintained in Eagle’s MEM (U87, SKNSH), DMEM (A172, HEK293T), or L-15 (SW620) media (Invitrogen), all with 10% Fetal Bovine Serum (Hyclone). Cells were transfected with PEI or Turbofect (Fisher) according to the manufacturer’s instructions. Protein expression was analyzed by western blot with antibodies to the Flag (Sigma, Flag M2) or V5 epitope tag (Cell Signaling, 13202). Expression of YAP1 was monitored using a YAP/TAZ specific antibody from Santa Cruz (SC-101119), and HSP90 was detected with an antibody from Cell Signaling (#4874). U87 and A172 cells were transduced with lentiviral vectors under standard conditions using polybrene, and 72 hours after infection cells were subjected to selection with 1μg/ml Puromycin or 2μg/ml Blasticidin, or 0.5μg/ml Puromycin plus 2μg/ml Blasticidin. Assays on infected cells were performed after at least 10 days in selection. A172 and U87 cells were authenticated by STR profiling, in accordance with ICLAC guidelines, at the University of Arizona Genetics Core.
DNA constructs
Myt1 and Myt1l constructs were generated by PCR from plasmids obtained from Addgene (pMycMyt1–7zf/IRES-Red, #22652, a gift from Lynn Hudson [43], and Tet-OFUW-Myt1l, #27152, a gift from Marius Wernig [18]). Lentiviral Myt1 and Myt1l expression constructs were generated in a modified pLenti puro (Addgene #39481, a gift from Le-Ming Shih [44]), into which we first inserted an amino-terminal triple Flag-tag [11]. YAP1-V5 and LacZ in pLX304 were gifts from William Hahn (Addgene #42555 and #42560 [45]). 8×GTIIC-luc was a gift from Stefano Piccolo (Addgene #34615 [46]), and pcDNA Flag Yap1 from Yosef Shaul (Addgene #18881 [47]). The shYAP1 viral plasmid, shYAP1 # 1, and the control shLacZ, were gifts from William Hahn (Addgene plasmids # 42540 and # 42559 [45]).
RNA and qRT-PCR
RNA was isolated and purified using Absolutely RNA kit (Agilent). For qRT-PCR, cDNA was generated using Superscript III (Invitrogen), and analyzed in triplicate by real time PCR using a BioRad MyIQ cycler and Sensimix Plus SYBRgreen plus FITC mix (Bioline). Intron spanning primers, selected using Primer3 (http://frodo.wi.mit.edu/), were used for all qRT-PCR. Expression is shown as mean plus standard deviation of triplicate experiments and was normalized to Rpl4 as described [48].
Proliferation assays
Cells were plated in four well chamber slides (Labtek2, Nunc) and incubated with EdU (ThermoFisher) for 45 minutes prior to fluorescent detection. pHH3 staining was performed as described [49], using an antibody from Millipore (#06–570). Long term proliferation assays were performed by serially plating 3×105 cells from independent quadruplicate cultures and counting cells after 3 or 4 days. For growth in soft agar, 5,000 or 10,000 cells per well were seeded into soft agar in 6-well plates [50]. Media on the colonies was changed every 4th day. After 21–28 days colonies, were stained with 0.005% crystal violet (Sigma-Aldrich) and imaged using a Gel Doc XR+ (BioRad). Colony number was counted using Quantity One 1-D Analysis Software (BioRad).
Luciferase assays
Cells were transfected with the 8×GTIIC-luc firefly luciferase reporter and a phCMVRLuc control (Promega, Madison, WI), with pcDNA Flag Yap1 as indicated, using PEI. After 48 hours, activity was assayed with luciferase assay reagent (Biotium) using a Berthold LB953 luminometer. Results were normalized using Renilla luciferase activity, assayed with 0.09μM coelenterazine (Biosynth, Naperville, IL), as in [51]. Results of replicate transfections are shown (N=3–6, mean plus standard deviation) normalized to the RLuc transfection control. The YAP1-luc reporter was a kind gift from Dr. R. Janknecht (University of Oklahoma) [52], and mutant versions were generated within this plasmid by PCR.
Chromatin immunoprecipitation (ChIP)
ChIP was performed on U87 cells following lentiviral transduction with Flag-tagged Myt1 or Myt1l, or with a control lentiviral vector. Chromatin was cross-linked for 20 minutes in 1% formaldehyde and sonicated to 200–1000bp using a Branson digital sonifier, with microtip as described [53]. Immunoprecipitation was carried out using 10μl of anti-Flag agarose or anti-GluGlu agarose as a negative control. Bound and input fractions were analyzed by qPCR on a BioRad MyIQ cycler using Sensimix Plus SYBRgreen plus FITC mix (Bioline).
Statistics and analysis of gene expression datasets
Comparison of qRT-PCR, proliferation and luciferase data was performed by Student’s T-test, with multiple correction testing where appropriate. Overlaps between RNA-seq data and GBM datasets were analyzed using a 2×2 contingency table and Chi-squared test. For comparison of gene expression data to expression correlations in GBM and LGG, we overlapped genes that were significantly increased or decreased (p-adj < 0.0001, log2 fc +/−0.5) in both Myt1 and Myt1l expressing U87 compared to control, with the top ~1000 negatively and positively correlated genes (using the minimum Spearman cut-off that yielded >1000 genes) from TCGA data. Relative expression in GBM and LGG datasets was compared relative to mean expression for all samples, and was plotted as median, with upper and lower quartiles (box) and 5th and 95th percentiles (whiskers). p-values were generated using a Student’s T-test. TCGA data for GBM and LGG is from [4, 54], and other gene expression data was from GEO datasets GSE4290 and GSE7696. RNA-seq data for Myt1 and Myt1l expressing U87 cells is from GSE103790 [11]. RNA-seq data was analyzed for GO enrichment (david.ncifcrf.gov; [55, 56]), and by GSEA (broadinstitute.org; [57, 58]). Survival analyses (Kaplan-Meier curves and log rank p-values) were performed at cBioPortal (cbioportal.org) or REMBRANDT (betastasis.com). Correlation between YAP1 and Myt expression was performed on expression z-scores using the full data-sets to determine the R squared coefficient of determination. For the combined Myt1+Myt1l, we averaged the z-score for each.
Xenograft assays
All animal procedures were approved by the Animal Care and Use Committee of the University of Virginia, which is fully accredited by the AAALAC. U87 cells infected with a lentiviral shYAP1 or control lentivirus were used after 14 days of Puromycin selection. Six week old athymic Foxn1 nude mice (Envigo) were used for orthotopic implantation with 3×105 cells per animal to generate xenografts, using eight animals per group. Three weeks after injection, mice were euthanized and analyzed by contrast enhanced T1-weighted MRI using a 7 Tesla Bruker/Siemens ClinScan small animal MRI at the UVA Molecular Imaging Core.
RESULTS
Myt1 and Myt1l limit proliferation of GBM cell lines.
We have previously examined gene expression changes in U87 glioblastoma cells in which we stably re-expressed either Myt1 or Myt1l [11]. This analysis suggested that both proteins have largely similar effects on gene expression, despite the potential for Myt1l but not Myt1 to activate transcription from synthetic reporters. While analyzing these U87 cells, we noticed that, compared to the control cells those expressing either Myt1 or Myt1l appeared to proliferate more slowly. Analysis of the gene expression changes in common between the Myt1 and Myt1l expressing cells compared to controls suggested a significant enrichment for GO terms associated with proliferation among the genes that decreased compared to control (Supplementary Fig 1A). In contrast, neuron development and differentiation GO terms were enriched in the gene set that increased in the Myt1 and Myt1l expressing cells (Supplementary Fig 1B). GSEA analysis supported this, with enrichment for chromosome segregation and E2F target genes in the control cells (Supplementary Fig 1C). In contrast, the Myt1 and Myt1l expressing cells were enriched for gene sets associated with synaptic signaling and neuronal function (Supplementary Fig 1D). In addition, there was an enrichment for an EMT signature in the control cells and cell adhesion in Myt expressing cells, suggesting effects on both proliferation and cell-cell contact.
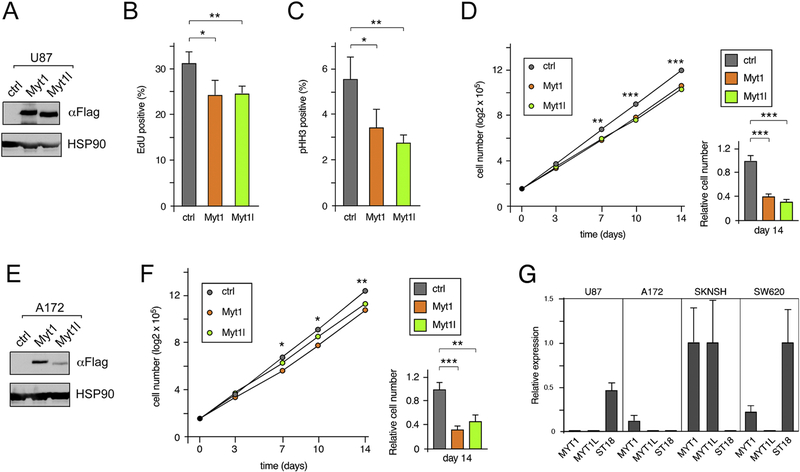
To test effects on proliferation, we first analyzed incorporation of EdU and examined the proportion of cells in late G2 or mitosis by staining for phospho-histone H3 (pHH3). As shown in Figure 1 (A-C), U87 cells expressing either Myt1 or Myt1l had small but significant reductions in the number of EdU and pHH3 positive cells. Serially passaging cells with 300,000 cells re-plated every 3 or 4 days resulted in a significant reduction in cumulative cell number by the second passage, with relative cell numbers below 40% of the control by the fourth passage (Fig 1D). Introduction of Myt1 or Myt1l into a second GBM cell line, A172, similarly reduced proliferation when assayed in a serial re-plating assay (Fig 1E, F). Analysis of U87 cells by qRT-PCR indicated that they do not express detectable levels of MYT1 or MYT1L, but have some expression of the related ST18 (Fig 1G). A172 cells expressed low levels of MYT1, but did not have detectable MYT1L or ST18 expression. In contrast, both MYT1 and MYT1L were readily detectable in the SKNSH neuroblastoma cell line, and ST18 expression was present in SW620 colon cancer cells. Thus U87 and A172 are good models for analyzing the function of reintroduced MYT proteins. This analysis suggests that reintroduction of either Myt1 or Myt1l can limit proliferation of glioblastoma cells in vitro and is consistent with a previous report that MYT1L inhibits GBM [20].
Figure 1. Decreased proliferation in Myt1 and Myt1l expressing cells.
A) Expression of Myt1 and Myt1l in U87 cells was analyzed by western blot with a Flag antibody (to the amino-terminal 3×Flag epitope). Blotting for HSP90 is shown below. B) U87 cells were incubated with EdU and the percentage of positive cells in quadruplicate cultures is plotted (mean + sd). C) The percentage of U87 cells in late G2 and M phase (mean + sd, of quadruplicates) was determined by staining with an antibody against pHH3. D) Four separate cultures of each U87 cell line were maintained for four serial passages, and the average cumulative cell number is plotted (left). The right hand panel shows relative cell number (mean + sd) after four passages. E) A172 cells were infected with control or Myt1 or Myt1l expressing lentiviral constructs. Expression of Myt1 and Myt1l in A172 cells was analyzed by Flag western blot, and blotting for HSP90 is shown below. F) Four separate cultures of each A172 line were maintained for four serial passages, and the average cumulative cell number is plotted (left). The right hand panel shows relative cell number (mean + sd) after four passages. p-values were determined by Student’s T-test: * p < 0.05, ** p < 0.01, *** p < 0.001. Panels B, C, D and F show representative experiments, with averages of quadruplicate samples from a single experiment. Panels B, C and F were repeated twice and panel D four times, with similar results. A and E are representative western blots (of three repeats), which were from the cells used in D and F. G) Relative expression of Myt family members in U87 and A172 is shown compared to SKNSH and SW620 as positive controls.
Expression of MYT1 or MYT1L correlates with survival in brain cancer.
Since expression of either Myt1 or Myt1l in U87 GBM cells results in similar changes in gene expression and reduced proliferation, we examined whether MYT1 as well as MYT1L expression levels correlated with tumor progression in human gene expression datasets. Comparison of RNA-seq data from Myt1 and Myt1l expressing U87 cells [11] with gene expression datasets from GBM patients [59, 60] revealed that among the genes that showed differences in our analysis and were different between normal and tumor tissue, the majority were increased by Myt1 or Myt1l expression and decreased in GBM tumors compared to normal (Supplementary Fig 2). This is consistent with the hypothesis that reduced expression of both MYT1 and MYT1L correlates with GBM progression.
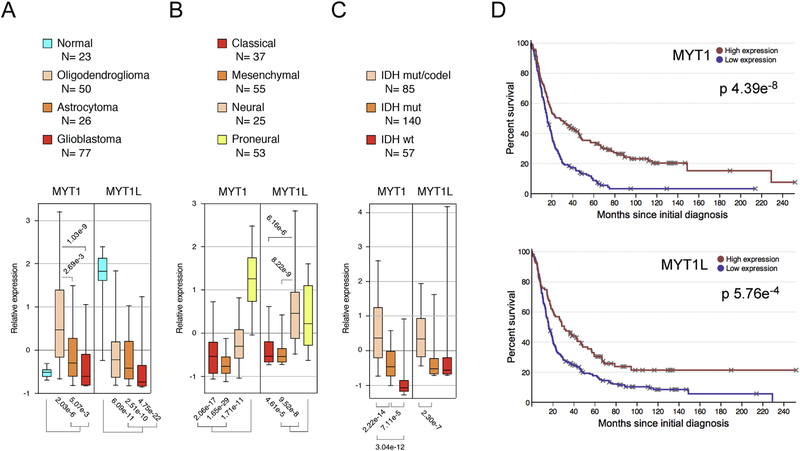
Examination of relative expression levels of MYT1 and MYT1L in human brain cancer datasets revealed significantly lower expression of MYT1L in oligodendroglioma, astrocytoma (grade III), and glioblastoma (grade IV) than in normal brain (Fig 2A). In contrast, MYT1 levels were higher in oligodendroglioma and astrocytoma than in normal, but expression in both astrocytoma and glioblastoma was significantly lower than in oligodendroglioma. Recent detailed molecular analyses of GBM and LGG patient samples have revealed new ways to classify these diseases that may be more informative than other histological classifications [4, 54]. Gene expression analysis separated GBMs into classical, mesenchymal, pro-neural, and neural subtypes, each with differing molecular signatures and clinical outcomes. Expression of both MYT1 and MYT1L was significantly higher in proneural tumors than in the more severe classical and mesenchymal tumors (Fig 2B). LGG has generally been classified by grade (II or III) and by histological type (oligodendroglioma and astrocytoma), but recent analysis suggests that three sub-types defined by mutations in IDH1/2 and deletion of 1p/19q may be more valuable. Analysis of TCGA data from LGG revealed higher expression of both MYT1 and MYT1L in the less aggressive subtype, characterized by the presence of IDH mutations and co-deletion of 1p and 19q chromosome arms (Fig 2C).
Figure 2. MYT1 and MYT1L expression in brain cancer datasets.
A) Relative expression of MYT1 and MYT1L in normal tissue, and three classifications of glioma is shown (data from GSE4290). Expression of MYT1 and MYT1L in the four subtypes of GBM (B) and in the three LGG subtypes (C) is shown (data from TCGA). Expression data is plotted as median with upper and lower quartiles (box) and 5th and 95th percentiles (whiskers). Numbers of samples in each class are indicated below. D) Disease-free survival is plotted for TCGA GBM data (from Betastasis) for patients with high or low MYT1 and MYT1L expression.
Analysis of survival data (REMBRANDT) revealed a significant association of higher expression of either MYT1 or MYT1L with longer survival of glioma patients (Fig 2D). Similarly, expression of either MYT1 or MYT1L was predictive of overall or diseasefree survival in the TCGA LGG dataset (Supplementary Fig 3A). Together these analyses suggest that both MYT1 and MYT1L expression correlates with the aggressiveness of human brain cancers, with expression being lower in more aggressive tumors and higher expression correlating with better overall survival.
Myt1 and Myt1l limit YAP1 expression.
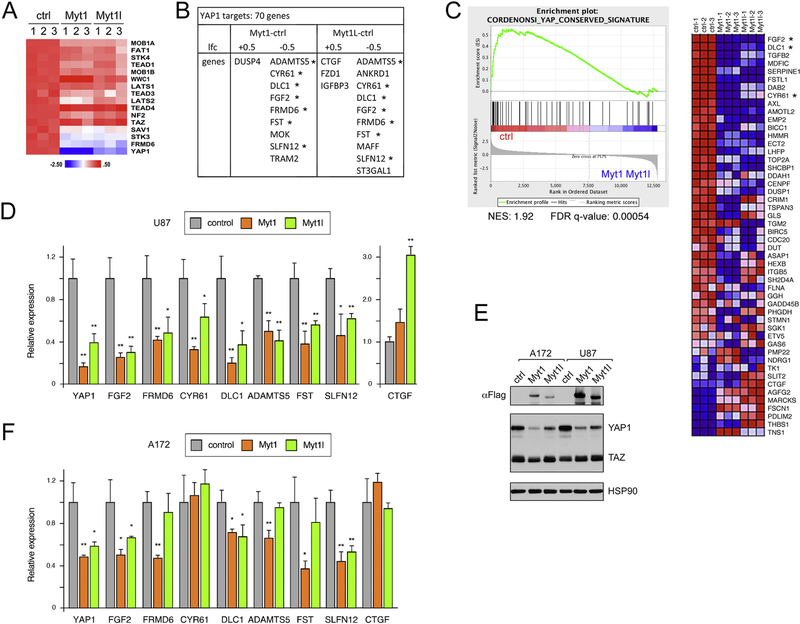
Examination of our gene expression data from U87 cells expressing either Myt1 or Myt1l suggested effects on the Hippo pathway, which was also the most enriched pathway by KEGG analysis among genes down-regulated by Myt1 and Myt1l (see Supplementary Fig 1A). This KEGG pathway includes the core components of the Hippo signaling pathway as well as target genes and additional regulatory inputs. Examination of expression levels of only the core components of the Hippo signaling pathway revealed that expression of YAP1 was the most clearly down-regulated in cells expressing either Myt1 or Myt1l, whereas the majority of rest of the core pathway was unchanged (Fig 3A). We overlapped our gene expression data with high confidence YAP1 targets, identified by combining YAP1 and TEAD1 ChIP-seq data with RNA-seq analysis of YAP1 knockdown from SF268 astrocytoma cells [61]. This revealed that more YAP1 targets are repressed than are activated by expression of Myt1 or Myt1l (Fig 3B). Additionally, by GSEA we found enrichment for higher expression of a conserved YAP1 signature [62] in the control data compared to cells expressing Myt1 or Myt1l (Fig 3C).
Figure 3. Regulation of YAP1 by Myt1 and Myt1l.
A) A heat map (z-score per gene) of RNA-seq data from Myt1 or Myt1l expressing and control U87 cells is shown for genes encoding components of the Hippo signaling pathway. B) Comparison of expression of high-confidence YAP1 target genes with changes in expression in U87 RNA-seq data, either +/− 0.5 log2 fold change (lfc). Genes with decreased expression in both Myt1 and Myt1l cells that were tested by qRT-PCR are indicated by asterisks. C) GSEA analysis of U87 RNA-seq data showing enrichment of a conserved YAP1 signature in the control cells. Three of the genes tested in D are near the top of this list (asterisks). D) qRT-PCR expression analysis for YAP1 and putative target genes (from panel B) in control and Myt1 or Myt1l expressing U87 cells. E) Western blot analysis (representative blot of three repeats) for Myt1, Myt1l, and YAP1 in A172 and U87 cells (control or expressing Myt1 or Myt1l). HSP90 loading control is shown below. F) A172 cells expressing Myt1 or Myt1l were analyzed by qRT-PCR as in D. p-values were determined by Student’s Ttest: * p < 0.05, ** p < 0.01. D and F are average and s.d. of triplicate samples from a single experiment.
To verify these potential changes in the Hippo pathway, we tested expression of YAP1 and seven predicted high confidence target genes from the SF268 dataset. We observed significantly lower expression of all eight genes in cells expressing Myt1 or Myt1l than in control U87 cells (Fig 3D). Western blot analysis for YAP1 confirmed the reduction in expression in both U87 and A172 cells expressing Myt1 or Myt1l compared to the control cells, with little change in the functionally related TAZ (Fig 3E). In A172 we observed significantly lower expression of the YAP1 mRNA and lower expression the majority of the predicted target genes identified from U87 RNA-seq analysis (Fig 3F). Expression of CTGF, a known YAP1 target gene, was not decreased by Myt expression in either U87 or A172 cells, and increased significantly in Myt1l expressing U87 cells (Fig 3D), perhaps suggesting an independent mechanism of regulation. This analysis suggests that expression of either Myt1 or Myt1l in GBM cells modulates the activity of the Hippo pathway by repressing expression of YAP1, and this reduces expression of at least some YAP1 target genes.
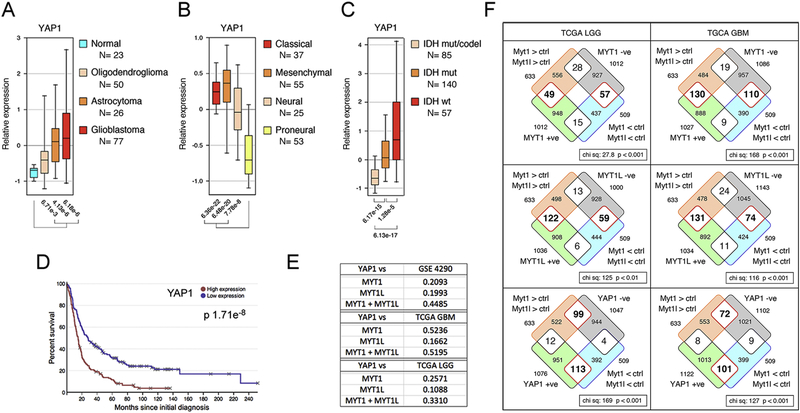
When we analyzed YAP1 expression in GBM subtypes we found an inverse pattern to that seen with MYT1 or MYT1L, with higher YAP1 expression in the all tumor types than in normal, and higher expression in the more aggressive classical and mesenchymal sub-types (Fig 4A, B). Similarly, relative YAP1 expression was lower in the less aggressive IDH mutant, 1p/19q co-deleted LGG subtype (Fig 4C). In contrast to MYT1 and MTY1L, high YAP1 expression was associated with poor survival in human glioma (Fig 4D). Similarly, in LGG decreased overall and disease-free survival correlated with higher YAP1 expression (Supplementary Fig 3B). Together with our gene expression analysis, these data suggest a potential inverse correlation between the expression of YAP1 and either MYT1 or MYT1L. Examination of expression patterns in normal human tissue using GTEx (gtexportal.org) supported this inverse correlation, with YAP1 expression being lower throughout the brain than in most other tissues, and MYT1L and MYT1 expression being higher in the brain (Supplementary Fig 4). Analysis of expression in the GBM and LGG datasets revealed a negative correlation between YAP1 and MYT1 expression (Fig 4E and Supplementary Fig 5). When we combined expression of both MYT1 and MYT1L, the correlation with YAP1 expression improved in one of the GBM datasets and in LGG.
Figure 4. Expression of YAP1 in brain cancers.
YAP1 expression was analyzed as in Figure 2, in normal and glioma from GSE4290 (A), TCGA GBM data (B), and LGG from TCGA (C). Numbers of samples in each class are indicated below. D) Kaplan-Meier analysis of YAP1 in TCGA GBM data (Betastasis) is shown. E) Correlation between zscores for expression of YAP1 and MYT1, MYT1L, or both together was analyzed in the indicated datasets. R-squared values are shown. F) Gene expression changes (Myt1 and Myt1l-expressing versus control cells) were compared to genes with expression that best correlated (either positively or negatively) with MYT1, MYT1L, or YAP1 expression in TCGA LGG, and GBM data-sets. The overlaps are shown as Venn diagrams, with the p-value (Chi squared test on a 2×2 contingency table) for the distribution of the overlap. Red outlines indicate the enriched overlap in each case.
To compare downstream gene expression changes to human cancer data-sets we identified the genes for which expression best correlated with MYT1, MYT1L, or YAP1 in TCGA LGG and GBM data-sets, taking approximately 1000 genes each with the best positive and negative correlation. These were then overlapped with genes that were significantly different between control U87 cells and those expressing Myt1 and Myt1l. As shown in Figure 4F, there was a preferential enrichment in the overlap for genes that were more highly expressed in Myt1 or Myt1l expressing U87 cells and positively correlated with their expression in either GBM or LGG. The converse was also true, enrichment for lower expression and a negative correlation, as might be expected, given that the U87 gene expression data is from cells expressing Myt1 or Myt1l. More interestingly, we observed the opposite pattern of enrichment among the overlaps between the Myt1/Myt1l U87 expression data and genes that correlated with YAP1 expression in GBM data sets (Fig 4F), consistent with decreased YAP1 mediating part of the effect of Myt1 or Myt1l in glioma. Taken together these analyses suggest that Myt1 and Myt1l limit YAP1 expression and that there is an inverse relationship between MYT1/MYT1L and YAP1 expression and function both in normal human tissue and in brain cancers.
Direct repression of YAP1 expression by MYTs
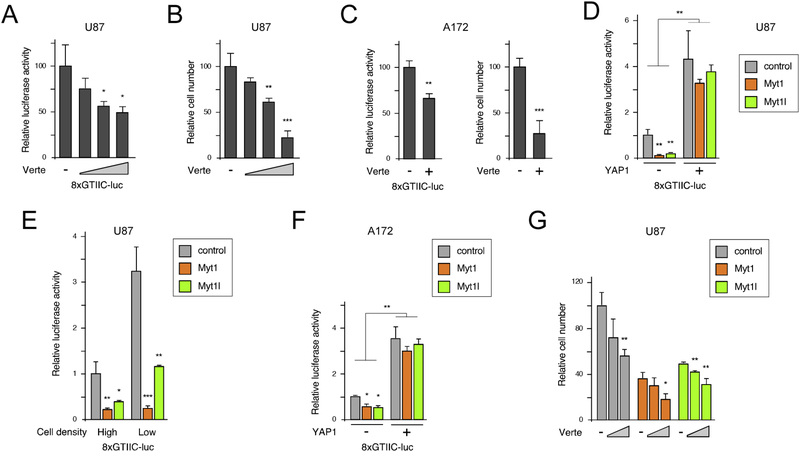
The effect of Myt1 and Myt1l expression on YAP1 prompted us to examine if the effects of Myt1 and Myt1l on proliferation in U87 cells might be due to decreased YAP1 levels. Verteporfin has been used to limit activation of YAP1 target genes by disrupting its interaction with the TEAD transcription factors, which mediate the majority of YAP1 recruitment to DNA [63]. To test if U87 cells were sensitive to modulation of this pathway, we treated cells with a range of concentrations of Verteporfin and examined the activity of a synthetic YAP1 transcriptional reporter (8×GTIIC-luc; [46]). As shown in Figure 5A, increasing Verteporfin decreased activation of this reporter in parental U87 cells, and also reduced total cell number when U87 cells were incubated with increasing concentrations of Verteporfin for four days (Fig 5B). Similarly, decreased reporter activity and cell number were seen in A172 cells treated with Verteporfin (Fig 5C).
Figure 5. GBM cell lines are sensitive to Hippo pathway modulation.
A) U87 cells were treated with increasing concentrations of Verteporfin (0, 2, 4, and 8 μM) after transfection with a YAP-TEAD luciferase reporter (8×GTIIC-luc) and assayed 24 hours later. B) U87 cells were incubated for four days with 0, 2, 4, or 8 μM Verteporfin, and relative cell number determined by counting quadruplicate cultures. C) A172 cells were analyzed as in A and B with 0 or 8 μM Verteporfin. D) U87 cells expressing Myt1 or Myt1l or with the control vector were transiently transfected with the 8×GTIIC-luc reporter with or without a YAP1 expression plasmid, and luciferase activity was determined after 48 hours. E) Cells transfected with the 8×GTIIC-luc reporter and were split at high and low density and assayed for luciferase activity 36 hours later. F) A172 cells expressing Myt1 or Myt1l or with the control vector were transfected with 8×GTIIC-luc with or without a YAP1 plasmid, and luciferase activity was determined after 48 hours. G) U87 cells expressing either Myt1 or Myt1l and control cells were incubated for four days with 0, 2, or 4 μM Verteporfin and relative cell number determined by counting quadruplicate cultures. All luciferase data is presented as mean + sd of triplicate transfections, normalized to a Renilla-luc transfection control. p-values were determined by Student’s T-test: * p < 0.05, ** p < 0.01, *** p < 0.001. Panels A-F show representative experiments, that were repeated twice with similar results. Data in panel G is from a single experiment. Averages shown are of triplicate samples for luciferase assays, and quadruplicates for proliferation assays, from a single representative experiment.
To test effects of Myt1 and Myt1l, we tested activity of the 8×GTIIC-luc YAP1 reporter in Myt1 or Myt1l expressing U87 cells. In both Myt1 and Myt1l expressing cells, YAP1 reporter activity was significantly lower than in the control cells, and this difference was abolished by overexpression of YAP1 (Fig 5D). Hippo signaling to YAP1 is sensitive to cell density, with more sparse cells allowing for increased YAP1 activation of target genes [46]. However, the effect of Myt1 and Myt1l appeared to be similar at both high and low density in U87 cells, consistent with decreased YAP1 expression in both cases (Fig 5E). As in U87, we observed lower 8×GTIIC-luc reporter activity in Myt1 and Myt1l expressing A172 cells that was overcome by YAP1 co-expression (Fig 5F). Examining cell number in cells treated with Verteporfin suggested that Myt1 or Myt1l expressing U87 cells retained some sensitivity to YAP1 inhibition, consistent with inhibition of residual YAP1 function by Verteporfin (Fig 5G). Together these results suggest that GBM cells are sensitive to inhibition of YAP1 activity, consistent with the hypothesis that reduced YAP1 levels may at least partly account for the effects of Myt1 and Myt1l reexpression on cell proliferation.
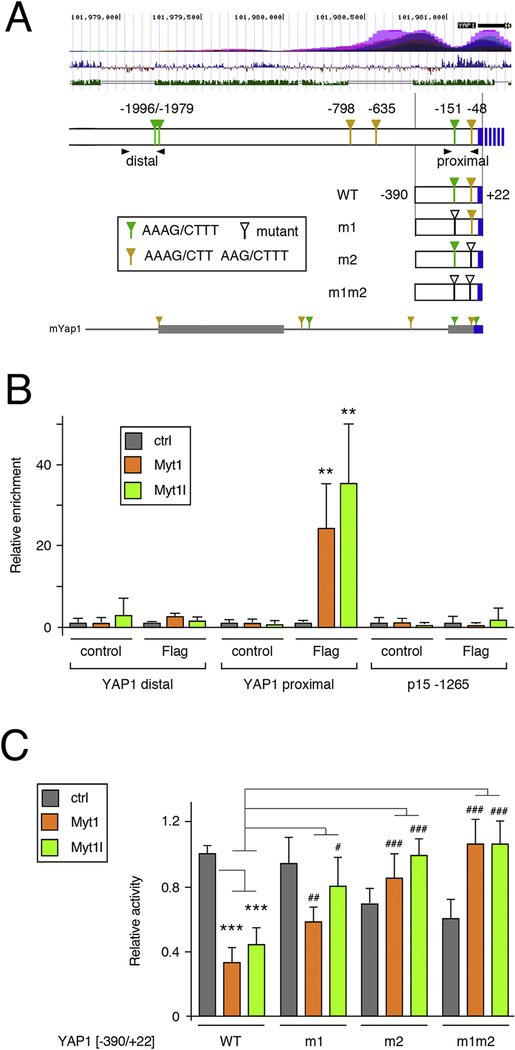
Examination of the YAP1 gene revealed multiple matches to the MYT consensus site within 2kb of sequence upstream of the TSS (Fig 6A). To test if Myt1 or Myt1l could bind to these sites we focused on the proximal promoter, since the potential MYT sites were conserved in mouse Yap1, and also tested a further upstream region with a closely spaced pair of sites, although these were not found in mouse Yap1 (Fig 6A). Binding of Myt1 or Myt1l from stably expressing U87 cells was analyzed by ChIP-qPCR. As shown in Fig 6B, we detected significant enrichment of both Myt1 and Myt1l at the proximal YAP1 promoter region, but no binding to the upstream region containing the paired sites, or to a control region from upstream of the CDNK2B gene. To determine if the consensus sites within this region of the YAP1 upstream mediated MYT-dependent repression we used a luciferase reporter that includes sequence to 390bp 5’ of the transcriptional start site [52]. Each of the two consensus sites present within this reporter was mutated individually (m1 or m2: Fig 6A), and we also generated the double mutant (m1m2) lacking both sites. Following transfection into U87 cells expressing either Myt1 or Myt1l we observed significantly lower activity from the wild type reporter than seen in control cells (Fig 6C). Mutating either site alone reduced repression by both Myt1 and Myt1l, and the activity of the double mutant was further increased, at least relative to the reporter with the m1 mutation alone. Together, these analyses suggest that YAP1 is a target for direct repression by Myt1 and Myt1l, and that this is mediated by binding to two consensus sites within the first 400bp upstream of the TSS.
Figure 6. Direct regulation of YAP1 by Myts.
A) The human YAP1 upstream region is shown schematically. Above is a screenshot from the USCS genome browser showing H3K27Ac ChIP-seq signal (top), vertebrate conservation (middle) and identity with mouse (lower track). The positions of the MYT consensus sites are shown, relative to the transcriptional start, together with the four luciferase constructs generated. Arrowheads indicate the positions of the PCR primer pairs for ChIP. Below is the region from the mouse Yap1 gene, with two blocks of high sequence identity shown as thicker bars. Consensus MYT sites in mYap1 are marked. B) Binding of Myt1 and Myt1l was analyzed by ChIP-qPCR from U87 cells with either Flag-Myt1, Flag-Myt1l or a control vector. Binding to the proximal and distal regions of the YAP1 promoter was analyzed, together with a negative control region from the CDKN2B gene. Average plus sd of quadruplicate IPs. p-values were determined by Student’s T-test: ** p < 0.01, for comparison to the IP from control cells. C) Luciferase activity of the four constructs shown in A was analyzed in U87 cells expressing Flag-Myt1, Flag-Myt1l or in control cells. p-values were determined by Student’s T-test, with correction for multiple testing: *** p < 0.001, for comparison of WT reporter activity to the control cells, # p < 0.05, ## p < 0.01, ### p < 0.001 for comparison of the mutant reporters to the WT in Myt1 or Myt1l expressing cells. For C, a single representative experiment is shown, from two repeats, with average and s.d. of six replicates. Data in panel B is from a single experiment, with average and s.d. of four replicates.
YAP1 promotes proliferation in GBM cells
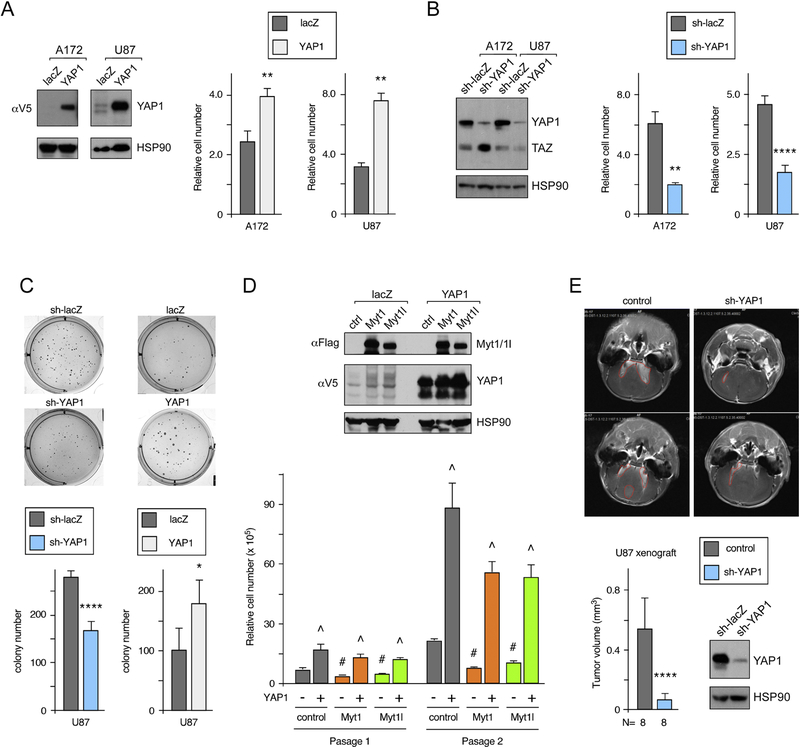
We next tested direct manipulation of YAP1 levels, either by overexpression or shRNA-mediated knockdown. As shown in Figure 7A, overexpression of YAP1 increased proliferation in both A172 and U87 cells. In contrast, knocking down YAP1 significantly reduced cell number in both lines (Fig 7B). In addition, altering YAP1 expression levels resulted in differing ability of U87 cells to form colonies in soft agar, with more colonies in the YAP1 overexpressing cells and fewer in cells in which endogenous YAP1 was decreased by shRNA knockdown (Fig 7C). Thus, expression of YAP1 in U87 or A172 cells has the opposite effect to that seen with expression of Myt1 or Myt1l. To test if the reduced proliferation in Myt1 or Myt1l expressing U87 cells could be reversed by over-expressing YAP1, we infected U87 cells with lentiviral constructs expressing either lacZ or YAP1, together with Myt1, Myt1l or the control vector. As shown in Figure 7D, expression of the epitope-tagged YAP1 did not appear to affect expression of Myt1 or Myt1l, compared to the lacZ expressing cells. Similarly, Myt1 or Myt1l expression did not reduce expression of the exogenous lentiviral YAP1. To examine effects on proliferation we performed a serial re-plating assay in which cells were counted after five days and then re-plated for a second passage. After only a single passage, and when the cumulative effects of both passages were considered, cells expressing Myt1 or Myt1l proliferated more slowly than controls, whereas YAP1 expressing cells proliferated at a significantly faster rate (Fig 7D). The combination of YAP1 and Myt1 or Myt1l largely reversed the anti-proliferative effect of Myt1 and Myt1l, although there was still some reduction compared to cells expressing YAP1 alone after two passages. These analyses show that Myt1/Myt1l and YAP1 have opposing effects on proliferation in GBM cell lines and suggest that reduced YAP1 expression in the presence of Myt1 or Myt1l may account for part of their anti-proliferative effect.
Figure 7. Analysis of proliferation with altered YAP1 expression.
A) U87 or A172 cells were infected with a control virus or a YAP1 expressing lentiviral construct. YAP1 expression was visualized by western blot for the V5 epitope tag. Quadruplicate cultures were counted after four days, and relative cell number is shown. B) YAP1 was knocked down by shRNA lentiviral infection in U87 or A172 cells. Endogenous YAP1 expression is shown by YAP1 western blot together with relative cell number after four days (quadruplicate cultures as in A). C) U87 cells with YAP1 overexpression or knockdown were assayed for growth in soft agar: 10,000 cells per well were plated for sh-lacZ and sh-YAP1, 5,000 cells per well for lacZ and YAP1. Average colony number and images of representative wells are shown. D) U87 cells were co-infected with lentiviral vectors expressing YAP1 (V5 epitope tagged) or lacZ and either the control Flag vector or one with 3×Flag Myt1 or Myt1l. Expression of Myt1, Myt1l and YAP1 was analyzed by western blot for the epitope tags, and HSP90 as a loading control is shown below. Quadruplicate cultures of each of the U87 lines were maintained for two serial passages and cell number after the first passage and cumulative cell number after two passages is shown (mean + sd). p-values were determined by Student’s T test (p < 0.01): # For Myt1 or Myt1 expressing lacZ versus control/lacZ, ^ for YAP1 expressing versus equivalent lacZ line. E) Control or shYAP1 infected U87 cells were injected orthotopically into nude mice and tumors imaged by MRI after three weeks. Average (+ s.d.) tumor volume is shown, together with representative images (red lines outline the tumors). For panels A-C and E, p-values were determined by Student’s T-test: * p < 0.05, ** p < 0.01, *** p < 0.001, *** p < 0.0001. Panels A, B and D show representative experiments that were repeated twice with similar results. The average and s.d. shown is based on replicates within a single experiment. Data for C and E are from single experiment, with average and s.d. shown based on replicates (6 wells each in C, and 8 mice each in E).
To test if YAP1 expression was required for tumor growth in vivo, we performed xenograft assays using U87 cells expressing a YAP1 shRNA, or a control vector. Cells were injected orthotopically into nude mice, and after three weeks, the tumors were imaged by MRI. As shown in Figure 7E, there was a significant reduction in tumor volume in mice injected with the shYAP1 cells compared to the control U87 cells (see also Supplementary Fig 6). This is clearly consistent with the model that YAP1 expression promotes glioma progression, and together with previous analyses, suggests that repression of YAP1 expression by Myt1 or Myt1l mediates at least part of their antitumorigenic effect.
DISCUSSION
Here we demonstrate that re-expression of Myt1 or Myt1l in GBM cell lines slows proliferation, that expression of both is lower in more aggressive sub-types of glioma, and that reduced expression correlates with poor prognosis in both GBM and LGG. Analysis of gene expression data suggests that Myts alter the output of the Hippo signaling pathway by directly repressing expression of the YAP1 coactivator. We show that compared to YAP1, Myt proteins have an opposing effect on proliferation of GBM cells, and that YAP1 promotes tumor growth in vivo.
Myt1l is perhaps best known as one of a cocktail of three transcription factors, together with Brn2 and Ascl1, that can reprogram fibroblasts to neurons [16–18]. In addition, Myt1, as well as the third family member, St18, can promote neuronal differentiation of P19 cells in vitro, suggesting similar roles for all three family members [15]. More recent analysis has suggested that Myt1l limits both non-neuronal gene expression programs and non-neuronal cell fate [10], consistent with the gene expression differences we observed between U87 cells and those with Myt1 or Myt1l reexpression. Analysis of gene expression patterns in adult human tissues (gtexportal.org) suggests that all three members of the family are primarily expressed in brain, with essentially no expression in other tissues, other than pituitary. In the developing mouse embryo, Myt1 and Myt1l are broadly expressed in neural tissues during the latter half of embryogenesis, with St18 expression being lower, but following a similar pattern [14]. Despite their neural-restricted expression, gene knock-out studies in mice suggest a role for Myt1, and possibly also St18, in the differentiation of endocrine islet cells and pancreatic function [64–66]. The similar expression patterns and function in neural development for Myt1 and Myt1l might predict similar mechanisms of transcriptional regulation, which is supported by the highly related DNA-binding zinc fingers, overall domain structure, and their preference for binding to the same DNA element [7–9, 67]. Initial analyses suggested that Myt1 and Myt1l might in fact have opposing effects on transcription when targeted to DNA via their cognate binding site [6, 8, 9, 12, 13]. However, direct comparison of the transcriptional consequences of re-expressing either Myt1 or Myt1l in U87 cells suggests that they perform largely overlapping functions [11]. This is further supported by our analysis here, showing similar effects on proliferation and the Hippo signaling pathway in two GBM cell lines.
Although the St18 member of this family was originally identified as a potential breast cancer tumor suppressor [19], the functions of this family of proteins have not been extensively studied in cancer. However, a role for MYT1L in GBM has been examined, with knock-down of MYT1L in neural stem cells resulting in increased tumorigenesis in an orthotopic tumor model [20]. Our data showing reduced proliferation with re-introduction of either Myt1 or Myt1l clearly support this, although we were unable to maintain expression of Myt1/Myt1l in U87 cells without selection, making testing in xenograft models problematic. Surprisingly, knock-down of Myt1 in the Neuro2a neuroblastoma cell line resulted in reduced proliferation, an effect that was also seen with knock-down of the Myt1-interacting LSD1 lysine demethylase [13]. Comparison of gene expression changes between Myt1 knock-down Neuro2a and Myt1-expressing U87 cells suggested that direct effects on gene expression were similar [11]. Thus it is possible that even in neural cells, the effects of this family of proteins on tumorigenesis may differ among tumor types, despite similar effects on target genes. In this context, it is of interest that cell line expression data from CCLE (betastasis.org) suggests that MYT1 and MYT1L expression is extremely low or absent in all CNS and neural derived tumor cell lines, other than in some neuroblastoma and medulloblastoma lines. Our analysis of MYT1 and MYT1L expression in human cancer data-sets supports a tumor-suppressive role in both LGG and GBM, with lower expression being seen in more aggressive tumor types. This is supported by in situ analysis for MYT1L transcripts, showing higher expression in normal brain and oligodendroglioma with 1p loss than in oligodendroglioma without LOH at 1p or in GBM [68]. One exception is that compared to normal tissue, MYT1 expression was higher in the less aggressive subtypes of GBM. However, expression of both MYT1 and MYT1L clearly decreases with increasing severity in both GBM and LGG, and low expression correlates with poor patient survival.
Examination of our gene expression data revealed reduced expression of the YAP1 coactivator that functions in the Hippo pathway, and a YAP1 expression signature was enriched in control cells compared to Myt1 or Myt1l expressing U87. In support of this we show that Myt1 and Myt1l regulate expression of YAP1 in both U87 and A172 cells, and that several YAP1 target genes are also suppressed in the presence of Myt1/Myt1l. Importantly, we show that repression of YAP1 by Myt1/Myt1l is mediated by direct binding to the proximal human YAP1 promoter, via two consensus MYT sites that also are conserved in mouse. In vitro analyses suggest binding of Myts to a closely spaced pair of sites and a so called DR9 element (direct repeat with 9bp spacing) has been widely used for functional analyses [9, 67]. Our previous analysis of gene expression changes with Myt1 or Myt1l expression suggested that the most repressed genes had multiple copies of the MYT consensus site close to the TSS [11]. However, we did not determine which sites were functional. Here we show that the Myt-responsive region of YAP1 contains two MYT consensus sites, separated by 99bp, both of which contribute to repression. However, we do not know if they are bound by a single Myt protein or each by distinct molecules, and it is possible that additional partial matches to the consensus may aid binding to each of these sites.
In addition to repression of YAP1 expression by Myt1 and Myt1l, genes with expression levels that best correlate with YAP1 in GBM or LGG are preferentially repressed by expression of Myt1 or Myt1l in U87 cells, consistent with decreased expression of YAP1 and its targets. YAP1, and the related TAZ, promote proliferation and survival in many cell types and have been implicated in a large number of cancers, where they can also contribute to metastasis and resistance to anti-cancer therapies [35, 69]. Previous work has suggested a role for YAP1 in promoting GBM [36, 37], and the functionally related TAZ has also been implicated [70, 71]. However, a link between the Myt transcription factors and expression of YAP1 has not been investigated. Analysis of human brain cancer data-sets suggests an inverse expression pattern comparing YAP1 with MYT1 and MYT1L, and survival data suggest that high YAP1 expression predicts poor survival in both GBM and LGG. This is again opposite to the results with MYT1 and MYT1L, further supporting antagonism between MYT1/MYT1L and YAP1 in glioma.
YAP1 and TAZ are transcriptional coactivators that function primarily via interaction with the TEAD family of DNA binding transcription factors [61, 72]. Upstream Hippo signaling exerts a negative regulatory function on YAP1 and TAZ [25]. Activation of this kinase cascade results in phosphorylation of YAP1/TAZ, which promotes cytoplasmic retention and degradation. While this regulatory module has been extensively studied, relatively little is known about transcriptional regulation of YAP1 or TAZ. Recent work has shown a role for the lysine demethylase, KDM4A, and Ets family DNA binding proteins, such as ETV1 in directly activating the YAP1 proximal promoter [52]. Activation of YAP1 expression by KDM4A and ETV1 promoted prostate cancer progression, suggesting that transcriptional up-regulation of YAP1 or TAZ can contribute to tumorigenesis. Our data provide a second example in which loss of transcriptional repression of YAP1 by MYT1/MYT1L may have a similar outcome, namely increased YAP1 expression driving tumor progression.
In summary, we show that Myt family zinc finger transcription factors directly repress YAP1 expression via two consensus sites close to the TSS, resulting in reduced proliferation in GBM cells. YAP1 and MYT1/MYT1L expression levels are inversely correlated in GBM and LGG and are predictive of survival. Decreasing endogenous YAP1 levels slows GBM proliferation and reduced tumor growth, suggesting that YAP1 activity might be viable target for treating glioma patients.
Supplementary Material
Highlights.
Myt zinc finger transcription factors directly repress expression of YAP1
Higher MYT1 and MYT1L expression correlates with better survival in GBM
High YAP1 expression correlates with poor survival in GBM and LGG
Reduced YAP1 expression slows GBM cell proliferation and tumor formation
Acknowledgements
The authors thank members of the Wotton lab for helpful discussions.
Funding
This work was supported by the National Institutes of Health/National Institute of Neurological Disorders and Stroke (grant number: NS077958).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Osuka S, Van Meir EG, Overcoming therapeutic resistance in glioblastoma: the way forward, J Clin Invest, 127 (2017) 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tanaka S, Louis DN, Curry WT, Batchelor TT, Dietrich J, Diagnostic and therapeutic avenues for glioblastoma: no longer a dead end?, Nat Rev Clin Oncol, 10 (2013) 14–26. [DOI] [PubMed] [Google Scholar]
- [3].Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO, R. European Organisation for, T. Treatment of Cancer Brain, G. Radiation Oncology, G. National Cancer Institute of Canada Clinical Trials, Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial, Lancet Oncol, 10 (2009) 459–466. [DOI] [PubMed] [Google Scholar]
- [4].Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L, The somatic genomic landscape of glioblastoma, Cell, 155 (2013) 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN, N. Cancer Genome Atlas Research, Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1, Cancer cell, 17 (2010) 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jiang Y, Yu VC, Buchholz F, O’Connell S, Rhodes SJ, Candeloro C, Xia YR, Lusis AJ, Rosenfeld MG, A novel family of Cys-Cys, His-Cys zinc finger transcription factors expressed in developing nervous system and pituitary gland, J Biol Chem, 271 (1996) 10723–10730. [DOI] [PubMed] [Google Scholar]
- [7].Kim JG, Hudson LD, Novel member of the zinc finger superfamily: A C2-HC finger that recognizes a glia-specific gene, Mol Cell Biol, 12 (1992) 5632–5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bellefroid EJ, Bourguignon C, Hollemann T, Ma Q, Anderson DJ, Kintner C, Pieler T, X-MyT1, a Xenopus C2HC-type zinc finger protein with a regulatory function in neuronal differentiation, Cell, 87 (1996) 1191–1202. [DOI] [PubMed] [Google Scholar]
- [9].Yee KS, Yu VC, Isolation and characterization of a novel member of the neural zinc finger factor/myelin transcription factor family with transcriptional repression activity, J Biol Chem, 273 (1998) 5366–5374. [DOI] [PubMed] [Google Scholar]
- [10].Mall M, Kareta MS, Chanda S, Ahlenius H, Perotti N, Zhou B, Grieder SD, Ge X, Drake S, Euong Ang C, Walker BM, Vierbuchen T, Fuentes DR, Brennecke P, Nitta KR, Jolma A, Steinmetz LM, Taipale J, Sudhof TC, Wernig M, Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates, Nature, 544 (2017) 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Manukyan A, Kowalczyk I, Melhuish TA, Lemiesz A, Wotton D, Analysis of transcriptional activity by the Myt1 and Myt1l transcription factors, Journal of cellular biochemistry, 119 (2018) 4644–4655. [DOI] [PubMed] [Google Scholar]
- [12].Romm E, Nielsen JA, Kim JG, Hudson LD, Myt1 family recruits histone deacetylase to regulate neural transcription, J Neurochem, 93 (2005) 1444–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yokoyama A, Igarashi K, Sato T, Takagi K, Otsuka IM, Shishido Y, Baba T, Ito R, Kanno J, Ohkawa Y, Morohashi K, Sugawara A, Identification of myelin transcription factor 1 (MyT1) as a subunit of the neural cell type-specific lysine-specific demethylase 1 (LSD1) complex, J Biol Chem, 289 (2014) 18152–18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Matsushita F, Kameyama T, Kadokawa Y, Marunouchi T, Spatiotemporal expression pattern of Myt/NZF family zinc finger transcription factors during mouse nervous system development, Dev Dyn, 243 (2014) 588–600. [DOI] [PubMed] [Google Scholar]
- [15].Kameyama T, Matsushita F, Kadokawa Y, Marunouchi T, Myt/NZF family transcription factors regulate neuronal differentiation of P19 cells, Neurosci Lett, 497 (2011) 74–79. [DOI] [PubMed] [Google Scholar]
- [16].Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, Wernig M, Induction of human neuronal cells by defined transcription factors, Nature, 476 (2011) 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Bjorklund A, Lindvall O, Jakobsson J, Parmar M, Direct conversion of human fibroblasts to dopaminergic neurons, Proc Natl Acad Sci U S A, 108 (2011) 10343–10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M, Direct conversion of fibroblasts to functional neurons by defined factors, Nature, 463 (2010) 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jandrig B, Seitz S, Hinzmann B, Arnold W, Micheel B, Koelble K, Siebert R, Schwartz A, Ruecker K, Schlag PM, Scherneck S, Rosenthal A, ST18 is a breast cancer tumor suppressor gene at human chromosome 8q11.2, Oncogene, 23 (2004) 9295–9302. [DOI] [PubMed] [Google Scholar]
- [20].Hu J, Ho AL, Yuan L, Hu B, Hua S, Hwang SS, Zhang J, Hu T, Zheng H, Gan B, Wu G, Wang YA, Chin L, DePinho RA, From the Cover: Neutralization of terminal differentiation in gliomagenesis, Proc Natl Acad Sci U S A, 110 (2013) 1452014527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lastowska M, Al-Afghani H, Al-Balool HH, Sheth H, Mercer E, Coxhead JM, Redfern CP, Peters H, Burt AD, Santibanez-Koref M, Bacon CM, Chesler L, Rust AG, Adams DJ, Williamson D, Clifford SC, Jackson MS, Identification of a neuronal transcription factor network involved in medulloblastoma development, Acta Neuropathol Commun, 1 (2013) 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ, The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation, Genes Dev, 9 (1995) 534–546. [DOI] [PubMed] [Google Scholar]
- [23].Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber D, Hariharan IK, salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines, Cell, 110 (2002) 467–478. [DOI] [PubMed] [Google Scholar]
- [24].Xu T, Wang W, Zhang S, Stewart RA, Yu W, Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase, Development, 121 (1995) 1053–1063. [DOI] [PubMed] [Google Scholar]
- [25].Yu FX, Guan KL, The Hippo pathway: regulators and regulations, Genes Dev, 27 (2013) 355–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML, TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm, Genes Dev, 15 (2001) 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, Sapkota G, Pan D, Massague J, Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways, Cell, 139 (2009) 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL, TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal, Nat Cell Biol, 10 (2008) 837–848. [DOI] [PubMed] [Google Scholar]
- [29].Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, Yaffe MB, TAZ: a novel transcriptional co-activator regulated by interactions with 14–3-3 and PDZ domain proteins, EMBO J, 19 (2000) 6778–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Oh H, Irvine KD, In vivo regulation of Yorkie phosphorylation and localization, Development, 135 (2008) 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ren F, Zhang L, Jiang J, Hippo signaling regulates Yorkie nuclear localization and activity through 14–3-3 dependent and independent mechanisms, Dev Biol, 337 (2010) 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, Li T, Chan SW, Lim CJ, Hong W, Zhao S, Xiong Y, Lei QY, Guan KL, The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase, J Biol Chem, 285 (2010) 37159–37169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhao B, Li L, Tumaneng K, Wang CY, Guan KL, A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP), Genes Dev, 24 (2010) 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Harvey KF, Zhang X, Thomas DM, The Hippo pathway and human cancer, Nat Rev Cancer, 13 (2013) 246–257. [DOI] [PubMed] [Google Scholar]
- [35].Zanconato F, Cordenonsi M, Piccolo S, YAP/TAZ at the Roots of Cancer, Cancer cell, 29 (2016) 783–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Orr BA, Bai H, Odia Y, Jain D, Anders RA, Eberhart CG, Yes-associated protein 1 is widely expressed in human brain tumors and promotes glioblastoma growth, J Neuropathol Exp Neurol, 70 (2011) 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Artinian N, Cloninger C, Holmes B, Benavides-Serrato A, Bashir T, Gera J, Phosphorylation of the Hippo Pathway Component AMOTL2 by the mTORC2 Kinase Promotes YAP Signaling, Resulting in Enhanced Glioblastoma Growth and Invasiveness, J Biol Chem, 290 (2015) 19387–19401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu M, Lin Y, Zhang XC, Tan YH, Yao YL, Tan J, Zhang X, Cui YH, Liu X, Wang Y, Bian XW, Phosphorylated mTOR and YAP serve as prognostic markers and therapeutic targets in gliomas, Lab Invest, 97 (2017) 1354–1363. [DOI] [PubMed] [Google Scholar]
- [39].Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL, Kim SH, Turski A, Azodi Y, Yang Y, Doucette T, Colman H, Sulman EP, Lang FF, Rao G, Copray S, Vaillant BD, Aldape KD, The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma, Genes Dev, 25 (2011) 2594–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jukam D, Xie B, Rister J, Terrell D, Charlton-Perkins M, Pistillo D, Gebelein B, Desplan C, Cook T, Opposite feedbacks in the Hippo pathway for growth control and neural fate, Science, 342 (2013) 1238016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lavado A, Ware M, Pare J, Cao X, The tumor suppressor Nf2 regulates corpus callosum development by inhibiting the transcriptional coactivator Yap, Development, 141 (2014) 4182–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang H, Deo M, Thompson RC, Uhler MD, Turner DL, Negative regulation of Yap during neuronal differentiation, Dev Biol, 361 (2012) 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nielsen JA, Berndt JA, Hudson LD, Armstrong RC, Myelin transcription factor 1 (Myt1) modulates the proliferation and differentiation of oligodendrocyte lineage cells, Mol Cell Neurosci, 25 (2004) 111–123. [DOI] [PubMed] [Google Scholar]
- [44].Guan B, Wang TL, Shih Ie M, ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers, Cancer Res, 71 (2011) 6718–6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, Shao DD, Schumacher SE, Weir BA, Vazquez F, Cowley GS, Root DE, Mesirov JP, Beroukhim R, Kuo CJ, Goessling W, Hahn WC, beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis, Cell, 151 (2012) 1457–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S, Role of YAP/TAZ in mechanotransduction, Nature, 474 (2011) 179–183. [DOI] [PubMed] [Google Scholar]
- [47].Levy D, Adamovich Y, Reuven N, Shaul Y, Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage, Mol Cell, 29 (2008) 350–361. [DOI] [PubMed] [Google Scholar]
- [48].Bjerke GA, Hyman-Walsh C, Wotton D, Cooperative transcriptional activation by Klf4, Meis2, and Pbx1, Mol Cell Biol, 31 (2011) 3723–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zerlanko BJ, Bartholin L, Melhuish TA, Wotton D, Premature senescence and increased TGFbeta signaling in the absence of Tgif1, PLoS ONE, 7 (2012) e35460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Carlton AL, Illendula A, Gao Y, Llaneza DC, Boulton A, Shah A, Rajewski RA, Landen CN, Wotton D, Bushweller JH, Small molecule inhibition of the CBFbeta/RUNX interaction decreases ovarian cancer growth and migration through alterations in genes related to epithelial-to-mesenchymal transition, Gynecol Oncol, 149 (2018) 350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hyman-Walsh C, Bjerke GA, Wotton D, An autoinhibitory effect of the homothorax domain of Meis2, The FEBS journal, 277 (2010) 2584–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kim TD, Jin F, Shin S, Oh S, Lightfoot SA, Grande JP, Johnson AJ, van Deursen JM, Wren JD, Janknecht R, Histone demethylase JMJD2A drives prostate tumorigenesis through transcription factor ETV1, J Clin Invest, 126 (2016) 706–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Anderson AE, Taniguchi K, Hao Y, Melhuish TA, Shah A, Turner SD, Sutherland AE, Wotton D, Tgif1 and Tgif2 Repress Expression of the RabGAP Evi5l, Mol Cell Biol, 37 (2017) e00527–00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].N. CancerGenome Atlas Research, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, Morozova O, Robertson AG, Noushmehr H, Laird PW, Cherniack AD, Akbani R, Huse JT, Ciriello G, Poisson LM, Barnholtz-Sloan JS, Berger MS, Brennan C, Colen RR, Colman H, Flanders AE, Giannini C, Grifford M, Iavarone A, Jain R, Joseph I, Kim J, Kasaian K, Mikkelsen T, Murray BA, O’Neill BP, Pachter L, Parsons DW, Sougnez C, Sulman EP, Vandenberg SR, Van Meir EG, von Deimling A, Zhang H, Crain D, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton T, Sherman M, Yena P, Black A, Bowen J, Dicostanzo K, Gastier-Foster J, Leraas KM, Lichtenberg TM, Pierson CR, Ramirez NC, Taylor C, Weaver S, Wise L, Zmuda E, Davidsen T, Demchok JA, Eley G, Ferguson ML, Hutter CM, Mills Shaw KR, Ozenberger BA, Sheth M, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Ayala B, Baboud J, Chudamani S, Jensen MA, Liu J, Pihl T, Raman R, Wan Y, Wu Y, Ally A, Auman JT, Balasundaram M, Balu S, Baylin SB, Beroukhim R, Bootwalla MS, Bowlby R, Bristow CA, Brooks D, Butterfield Y, Carlsen R, Carter S, Chin L, Chu A, Chuah E, Cibulskis K, Clarke A, Coetzee SG, Dhalla N, Fennell T, Fisher S, Gabriel S, Getz G, Gibbs R, Guin R, Hadjipanayis A, Hayes DN, Hinoue T, Hoadley K, Holt RA, Hoyle AP, Jefferys SR, Jones S, Jones CD, Kucherlapati R, Lai PH, Lander E, Lee S, Lichtenstein L, Ma Y, Maglinte DT, Mahadeshwar HS, Marra MA, Mayo M, Meng S, Meyerson ML, Mieczkowski PA, Moore RA, Mose LE, Mungall AJ, Pantazi A, Parfenov M, Park PJ, Parker JS, Perou CM, Protopopov A, Ren X, Roach J, Sabedot TS, Schein J, Schumacher SE, Seidman JG, Seth S, Shen H, Simons JV, Sipahimalani P, Soloway MG, Song X, Sun H, Tabak B, Tam A, Tan D, Tang J, Thiessen N, Triche T Jr., Van Den Berg DJ, Veluvolu U, Waring S, Weisenberger DJ, Wilkerson MD, Wong T, Wu J, Xi L, Xu AW, Yang L, Zack TI, Zhang J, Aksoy BA, Arachchi H, Benz C, Bernard B, Carlin D, Cho J, DiCara D, Frazer S, Fuller GN, Gao J, Gehlenborg N, Haussler D, Heiman DI, Iype L, Jacobsen A, Ju Z, Katzman S, Kim H, Knijnenburg T, Kreisberg RB, Lawrence MS, Lee W, Leinonen K, Lin P, Ling S, Liu W, Liu Y, Liu Y, Lu Y, Mills G, Ng S, Noble MS, Paull E, Rao A, Reynolds S, Saksena G, Sanborn Z, Sander C, Schultz N, Senbabaoglu Y, Shen R, Shmulevich I, Sinha R, Stuart J, Sumer SO, Sun Y, Tasman N, Taylor BS, Voet D, Weinhold N, Weinstein JN, Yang D, Yoshihara K, Zheng S, Zhang W, Zou L, Abel T, Sadeghi S, Cohen ML, Eschbacher J, Hattab EM, Raghunathan A, Schniederjan MJ, Aziz D, Barnett G, Barrett W, Bigner DD, Boice L, Brewer C, Calatozzolo C, Campos B, Carlotti CG Jr., Chan TA, Cuppini L, Curley E, Cuzzubbo S, Devine K, DiMeco F, Duell R, Elder JB, Fehrenbach A, Finocchiaro G, Friedman W, Fulop J, Gardner J, Hermes B, Herold-Mende C, Jungk C, Kendler A, Lehman NL, Lipp E, Liu O, Mandt R, McGraw M, McLendon R, McPherson C, Neder L, Nguyen P, Noss A, Nunziata R, Ostrom QT, Palmer C, Perin A, Pollo B, Potapov A, Potapova O, Rathmell WK, Rotin D, Scarpace L, Schilero C, Senecal K, Shimmel K, Shurkhay V, Sifri S, Singh R, Sloan AE, Smolenski K, Staugaitis SM, Steele R, Thorne L, Tirapelli DP, Unterberg A, Vallurupalli M, Wang Y, Warnick R, Williams F, Wolinsky Y, Bell S, Rosenberg M, Stewart C, Huang F, Grimsby JL, Radenbaugh AJ, Zhang J, Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas, N Engl J Med, 372 (2015) 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Huang da W, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nature protocols, 4 (2009) 44–57. [DOI] [PubMed] [Google Scholar]
- [56].Huang da W, Sherman BT, Lempicki RA, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists, Nucleic Acids Res, 37 (2009) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC, PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes, Nat Genet, 34 (2003) 267–273. [DOI] [PubMed] [Google Scholar]
- [58].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles, Proc Natl Acad Sci U S A, 102 (2005) 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, Hainfellner JA, Heppner FL, Dietrich PY, Zimmer Y, Cairncross JG, Janzer RC, Domany E, Delorenzi M, Stupp R, Hegi ME, Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma, J Clin Oncol, 26 (2008) 3015–3024. [DOI] [PubMed] [Google Scholar]
- [60].Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, Rosenblum M, Mikkelsen T, Fine HA, Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain, Cancer cell, 9 (2006) 287–300. [DOI] [PubMed] [Google Scholar]
- [61].Stein C, Bardet AF, Roma G, Bergling S, Clay I, Ruchti A, Agarinis C, Schmelzle T, Bouwmeester T, Schubeler D, Bauer A, YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers, PLoS genetics, 11 (2015) e1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, Daidone MG, Dupont S, Basso G, Bicciato S, Piccolo S, The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells, Cell, 147 (2011) 759–772. [DOI] [PubMed] [Google Scholar]
- [63].Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D, Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP, Genes Dev, 26 (2012) 1300–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Henry C, Close AF, Buteau J, A critical role for the neural zinc factor ST18 in pancreatic beta-cell apoptosis, J Biol Chem, 289 (2014) 8413–8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Tennant BR, Islam R, Kramer MM, Merkulova Y, Kiang RL, Whiting CJ, Hoffman BG, The transcription factor Myt3 acts as a pro-survival factor in beta-cells, PLoS ONE, 7 (2012) e51501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wang S, Zhang J, Zhao A, Hipkens S, Magnuson MA, Gu G, Loss of Myt1 function partially compromises endocrine islet cell differentiation and pancreatic physiological function in the mouse, Mech Dev, 124 (2007) 898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gamsjaeger R, O’Connell MR, Cubeddu L, Shepherd NE, Lowry JA, Kwan AH, Vandevenne M, Swanton MK, Matthews JM, Mackay JP, A structural analysis of DNA binding by myelin transcription factor 1 double zinc fingers, J Biol Chem, 288 (2013) 35180–35191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Mukasa A, Ueki K, Ge X, Ishikawa S, Ide T, Fujimaki T, Nishikawa R, Asai A, Kirino T, Aburatani H, Selective expression of a subset of neuronal genes in oligodendroglioma with chromosome 1p loss, Brain Pathol, 14 (2004) 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Moroishi T, Hansen CG, Guan KL, The emerging roles of YAP and TAZ in cancer, Nat Rev Cancer, 15 (2015) 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yang R, Wu Y, Wang M, Sun Z, Zou J, Zhang Y, Cui H, HDAC9 promotes glioblastoma growth via TAZ-mediated EGFR pathway activation, Oncotarget, 6 (2015) 7644–7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Yuan J, Xiao G, Peng G, Liu D, Wang Z, Liao Y, Liu Q, Wu M, Yuan X, MiRNA-125a-5p inhibits glioblastoma cell proliferation and promotes cell differentiation by targeting TAZ, Biochemical and biophysical research communications, 457 (2015) 171–176. [DOI] [PubMed] [Google Scholar]
- [72].Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, Piccolo S, Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth, Nat Cell Biol, 17 (2015) 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.