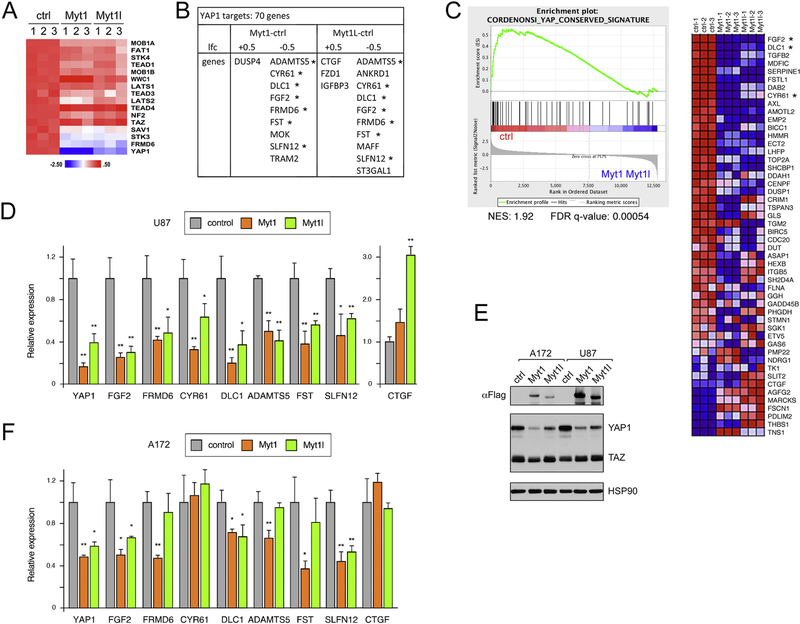
Figure 3. Regulation of YAP1 by Myt1 and Myt1l.
A) A heat map (z-score per gene) of RNA-seq data from Myt1 or Myt1l expressing and control U87 cells is shown for genes encoding components of the Hippo signaling pathway. B) Comparison of expression of high-confidence YAP1 target genes with changes in expression in U87 RNA-seq data, either +/− 0.5 log2 fold change (lfc). Genes with decreased expression in both Myt1 and Myt1l cells that were tested by qRT-PCR are indicated by asterisks. C) GSEA analysis of U87 RNA-seq data showing enrichment of a conserved YAP1 signature in the control cells. Three of the genes tested in D are near the top of this list (asterisks). D) qRT-PCR expression analysis for YAP1 and putative target genes (from panel B) in control and Myt1 or Myt1l expressing U87 cells. E) Western blot analysis (representative blot of three repeats) for Myt1, Myt1l, and YAP1 in A172 and U87 cells (control or expressing Myt1 or Myt1l). HSP90 loading control is shown below. F) A172 cells expressing Myt1 or Myt1l were analyzed by qRT-PCR as in D. p-values were determined by Student’s Ttest: * p < 0.05, ** p < 0.01. D and F are average and s.d. of triplicate samples from a single experiment.