Figure 1.
A gold standard of the cell-to-cell heterogeneity in DNA methylation
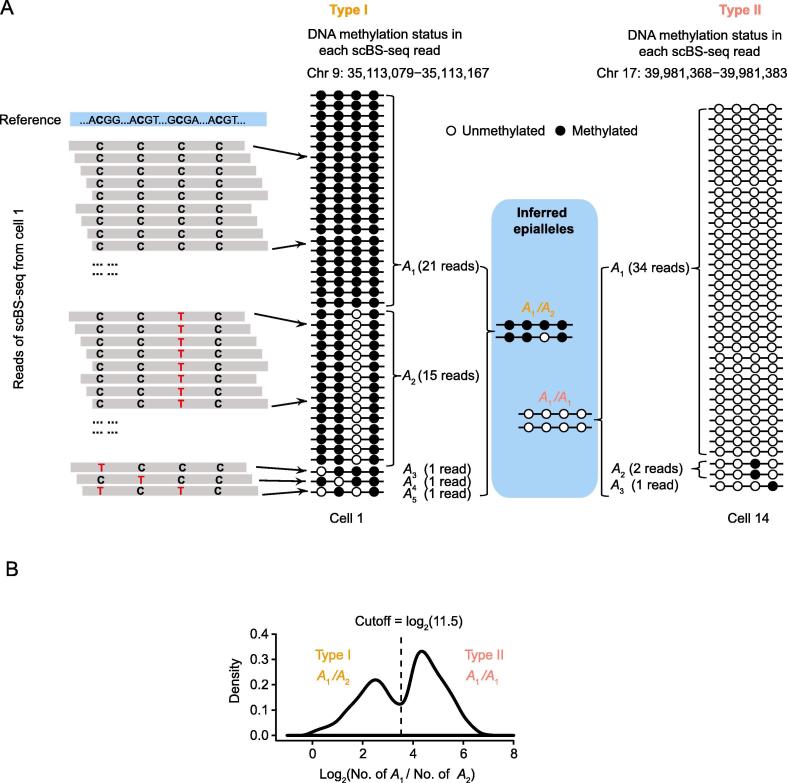
A. Representative scBS-seq reads from cell 1 that were mapped to the mouse genome (Chr 9: 35,113,079–35,113,167) are shaded. Cytosine in a BS-seq read represents a methylated CpG site, whereas thymine represents an unmethylated CpG site. Examples of heterozygous (A1/A2, Chr 9: 35,113,079–35,113,167 in cell 1) and homozygous (A1/A1, Chr 17: 39,981,368–39,981,383 in cell 14) epigenetic status are shown, respectively. The identity and number of all the methylation patterns in a cell are shown. Circles represent 4 consecutive CpG sites in a DNA segment, among which closed ones represent methylated sites and open ones represent unmethylated sites. Methylation patterns are ranked by their frequencies and Ai represents the ith methylation pattern. B. The distribution of the log2-transformed frequency ratio between the top 2 methylation patterns. The dashed line indicates the cutoff of the frequency ratio. If there is only one methylation pattern present in a DNA segment, a “pseudo” methylation pattern was added with a read count equal to 1.