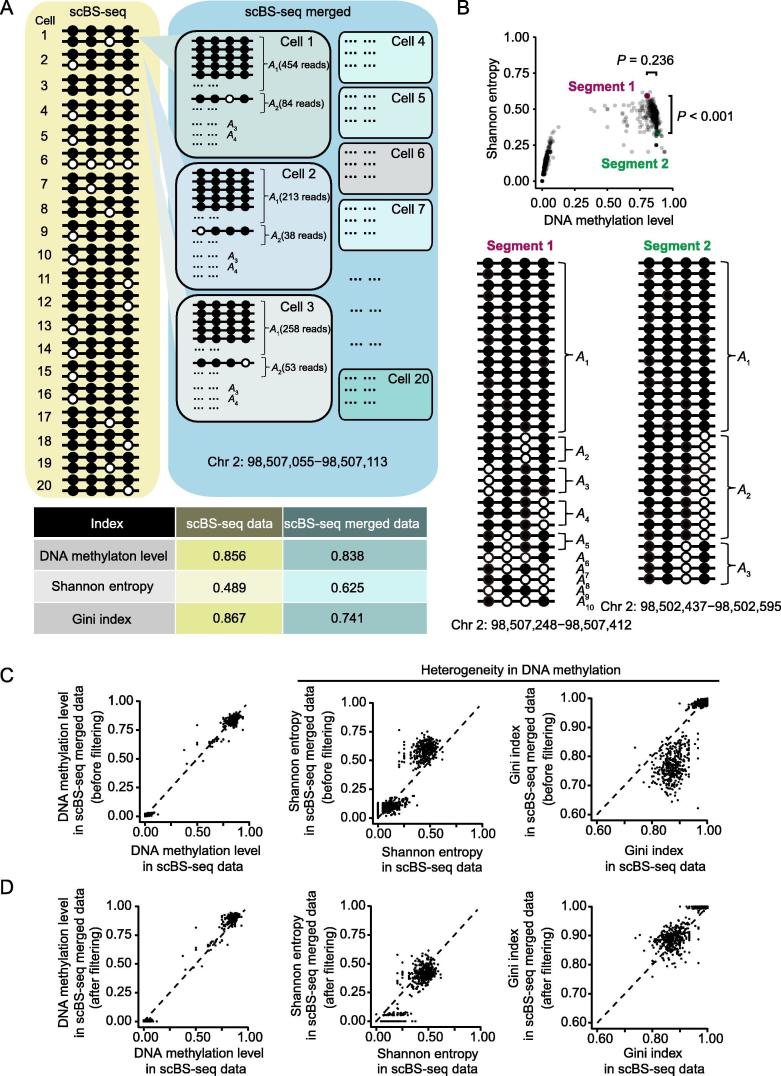
Figure 2.
Reproducing the gold standard from the in silico merged scBS-seq data
A. Heterogeneity in DNA methylation estimated from the 40 epialleles that were identified in the mouse scBS-seq data of a DNA segment (Chr 2: 98,507,055–98,507,113). DNA methylation level and heterogeneity (Shannon entropy and Gini index) of this segment are provided for both scBS-seq data (40 epialleles identified from 20 cells) and the in silico merged scBS-seq data (all sequencing reads from 20 cells), respectively. B. An example of two DNA segments (Chr 2: 98,507,248–98,507,412 and Chr 2: 98,502,437–98,507,595) that exhibit similar DNA methylation levels but exhibit different extents of heterogeneity. Each dot represents a DNA segment that contains 4 consecutive CpG sites. The epialleles identified in single cells are provided (the purple and green dots). Note that the DNA methylation status was not identified in every single cell due to the limited sequencing depth in scBS-seq. P values were calculated from the permutation test. C. Heterogeneity calculated from the unfiltered merged data. The dashed line represents y = x. Heterogeneity calculated from the merged data is overestimated (greater Shannon entropies and smaller Gini indices). D. The gold standard can be faithfully reproduced from the merged data with the filtered data.