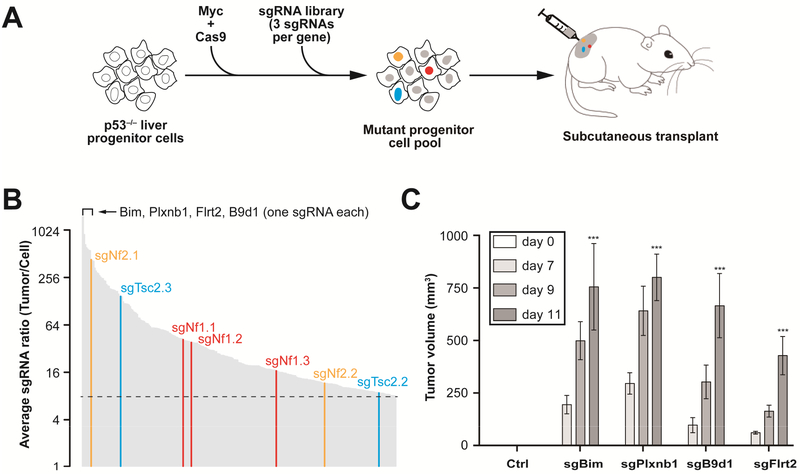
Figure 1. Genome-wide ex vivo CRISPR screen identifies new liver tumor suppressor genes.
(A) Outline of the screening strategy 8 sgRNAs targeting tumor suppressors accelerate formation of subcutaneous tumors and are enriched in the tumor.
(B) Average ratio of 267 individual sgRNAs enriched > 8-fold in tumors versus cell pool measured by high-throughput sequencing (n = 8). All three Nf1 sgRNAs (sgNf1.1, 2, 3) were enriched. Known liver tumor suppressors (Nf2, Tsc2) with two enriched sgRNAs, and new candidates (Bim, Plxnb1, etc) with one enriched sgRNA are highlighted.
(C) Validation of a subset of top-scoring sgRNAs in the subcutaneous tumor assay (n = 4 tumors). Average volume (n = 4) of tumors derived p53−/−; Myc; Cas9 cells infected with a control sgGFP (Ctrl) and a subset of top-scoring sgRNAs (sgBim, sgPlxnb1, sgB9d1, sgFlrt2). ***, P < .001. Error bars, mean ± s.e.m.