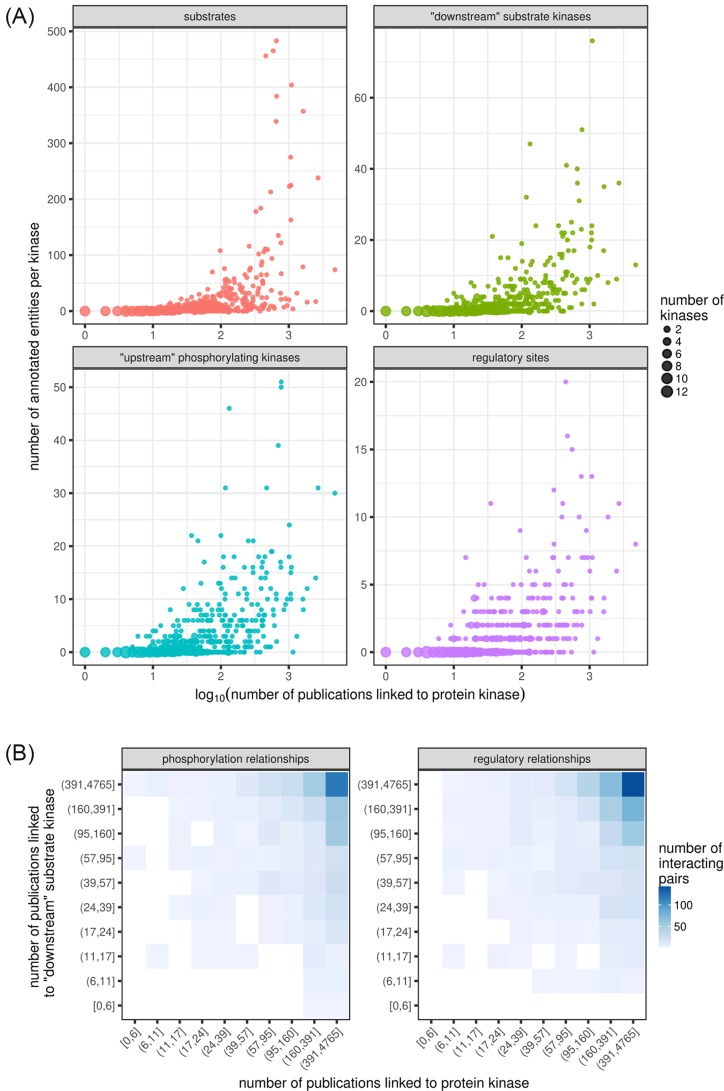
Figure 1. Study bias is a hindrance to comprehensive signalling pathway reconstruction.
(A) Well-studied human protein kinases may appear to be signalling hubs. They have more annotated substrates (top-left panel), including more substrates that are other protein kinases (top-right). They also are annotated to be substrates of more protein kinases themselves (bottom-left) and to carry more activity-regulating phosphorylation sites (bottom-right). Linked publications were retrieved from NCBI Entrez Gene database for each human protein kinase and filtered to remove publications associated with more than ten kinases [11]. Substrate and regulatory site counts were retrieved from PhosphoSitePlus [9]. All data were retrieved on 1 May 2018. (B) Literature-derived signalling networks are biased towards canonical pathways: annotated kinase-kinase phosphorylation (left) and regulatory (right) relationships tend to be between well-studied kinases, while relationships involving lesser studied kinases remain unknown. Publication counts for each kinase were binned into ten categories (deciles) of approximately 50 protein kinases each. Human kinase-kinase regulatory relationships were retrieved from OmniPath [10], restricting to directed relationships with support from at least two source databases. Autophosphorylation and autoregulation were omitted for clarity.