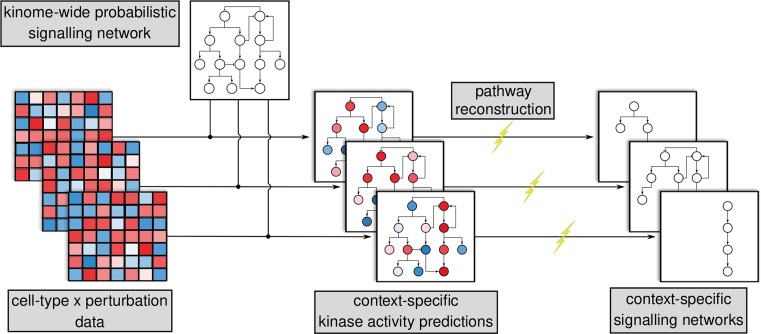
Figure 2. A kinome-wide, probabilistic regulatory network would serve as a foundation upon which to derive context-specific signalling pathways.
Phosphoproteomic and other data are collected from large-scale experiments in which different cellular contexts (e.g. cell lines, tissue types or stimulus regimes) are measured across several perturbations and/or time points. Kinase activities are inferred from these data and mapped on to the probabilistic network. Further network refinement is then performed to reconstruct context-specific signalling pathways.