Abstract
Ex vivo mucosal explants have become a mainstay of HIV-1 studies using human tissue. In this study, we examine the baseline phenotypic and virologic differences between biopsies derived from the small intestine (SI) and large intestine (LI) for use in ex vivo explant studies. To do this, we collected endoscopic mucosal biopsies from both SI and LI from the same healthy, HIV-seronegative participants. Mucosal mononuclear cell phenotypes and quantity were compared using flow cytometry. Comparative HIV-1 infectibility of the explants was assessed using an ex vivo explant HIV-1 infection assay. We found that all biopsies had similar numbers of T cells per biopsy. While the percentage of CD4+ T cells from SI biopsies expressed significantly more activation markers (CD38, HLA-DR) and HIV coreceptors (CXCR4, CCR5), the absolute numbers of activated CD4+ T cells were similar between both sites. LI explants, however, supported more efficient HIV-1 infection, as evidenced by earlier rise in p24 accumulation and greater percent of infected explants at limiting infectious doses. These results suggest that explants from LI biopsies support more efficient HIV-1 infection than SI biopsies, despite similar numbers of available, activated HIV-1 target cells. These findings highlight important differences in LI and SI explants, which must be considered in designing and interpreting ex vivo HIV-1 infection studies, and suggest that factors within the tissue other than target cell number and activation state may play a role in regulating HIV-1 infection.
Keywords: : HIV-1 infection, intestinal mucosa, large intestine, small intestine
Introduction
Mucosal surfaces of the genital and gastrointestinal (GI) tract are the predominant sites of HIV-1 transmission. Among sexual exposures, anal receptive intercourse is associated with the highest incidence of transmission, highlighting the unique susceptibility of the rectal mucosa.1 While the large intestine (LI) is the most commonly exposed site, the small intestine (SI) may be involved in vertical transmission via ingestion of infected amniotic fluid, blood, cervical secretions, or breast milk, as well as by secondary systemic spread following sexual exposure. Regardless of mode of transmission, the GI mucosa serves a primary site of HIV-1 replication in early infection.2–5
The human GI tract is in constant contact with antigens from both commensal and pathogenic organisms. The mucosal immune system must, therefore, be able to regulate a mounting, rapid, effective, immune response with tolerance. Regions of the GI tract can be classified based on the different absorptive and immune functions. The SI, which comprises the duodenum, the jejunum, and the ileum, engages in both secretory and absorptive functions within the digestive system. The LI, also referred to as the colon (including the ascending, transverse, descending, rectosigmoid, and rectal regions), houses the majority of the microbiome and performs a predominantly absorptive role in terms of digestion. The SI is lined with active immune cells in both the subepithelial and intraepithelial compartments, and the more distal SI contains numerous organized lymphoid regions termed Peyer's Patches (PP). Both SI and LIs have large numbers of activated, primarily memory lymphoid cells scattered throughout the lamina propria.
A practical consequence of this regional heterogeneity in structure and function is that translational observations concerning immunopathogenesis made in one region of the GI tract may not be valid for a more distant region. Indeed, a comparison of gene expression data throughout the intestine in HIV-infected persons showed regional differences between the SI and LI, with the SI more associated with inflammatory genes and LI more associated with changes in microbiome.6 Similarly, microbial communities differ among anatomic sites and are thought to, in part, influence the differential function.7
Given that the GI mucosa serves as a primary portal of entry for HIV-1, and is the major site of viral replication during primary infection regardless of mode of transmission, it is important to develop effective models to examine the role of the different mucosal regions in the early events of HIV-1 infection. Ex vivo explant models have become a primary model for studies of HIV-1 pathogenesis and early testing of therapeutics using human tissues.8–10 In this report, we present a comparative analysis of explants from the different mucosal regions (LI vs. SI) in terms of T cell composition, HIV-1 susceptibility, and sustained infection.
Materials and Methods
Ethics statement
This study was approved by the UCLA Office of the Human Research Protection Program Institutional Review Board (IRB) and all subjects signed written informed consent at the time of biopsy collection.
Study participants and tissue samples
HIV-seronegative participants (n = 7) were recruited from the UCLA Mucosal Immunology Core Laboratory registry (UCLA IRB No. 10-000528). Biopsy specimens were obtained endoscopically using a flexible sigmoidoscope and large cup radial jaw forceps (Microvasive Radial Jaw No. 1589, outside diameter 3.3 mm). The rectosigmoid colon was routinely sampled between 10 and 30 cm from the anal verge, whereas the SI was sampled in the second portion of the duodenum just above the ampulla of Vater. At each procedure, 16–18 biopsies were obtained from each site. Biopsies were placed immediately into 25 mL of RPMI 1640 medium supplemented with 10% fetal bovine serum, 2.5 mg/mL amphotericin B, and 0.1 mg/mL piperacillin–tazobactam and then transported to the laboratory within 1–2 h of collection.
Mucosal mononuclear cell phenotyping
Mucosal mononuclear cells (MMCs) were isolated from three to four pooled endoscopic biopsies for each individual participant using collagenase digestion as previously described.11 MMCs were stained with the following antibody panels (BD Biosciences, San Jose, CA) and then analyzed using flow cytometry. To enumerate total T cells, CD3-FITC (clone UCHT1), CD4-PE (clone SK3), CD45-PerCP (clone 2D1), and CD8-APC (cone SK1) were used; to enumerate activated T cells HLA-DR-FITC (clone L243), CD38-PE (clone HB7), CD4-PerCP (clone SK3), and CD8-APC (clone SK1) were used; to enumerate coreceptor expressing cells CD4-FITC (clone SK3), CCR5-PE (clone 2D7), CD45-PerCP (clone 2D1), and CXCR4-APC (clone 12G5) were used. All samples were acquired using a BD FACSCalibur flow cytometer (BD Biosciences). A minimum of 10,000 events were collected, and if this was not possible due to limited cell quantities then that data were excluded from further analysis. Analysis was performed using FlowJo (v10; FlowJo LLC). Cells were gated on live cells using forward- and side-scatter parameters, and then, lymphocytes were gated using CD45 and side-scatter parameters. Gating strategy is shown in Supplementary Figure S1 (Supplementary Data are available online at www.liebertpub.com/aid). For absolute cell number quantitation, MMC suspensions were stained with Trucount reagents as per the manufacturer's instructions (BD Biosciences).
HIV-1 virus expansion
The following reagent was obtained from the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH: HIV-1BaL from Dr. Suzanne Gartner, Dr. Mikulas Popovic, and Dr. Robert Gallo. Stocks of HIV-1BaL were prepared by infection using PM1 cells. TCID50 was determined by titration using pooled peripheral blood mononuclear cells. The same viral stock was used throughout the study.
HIV-1 infection of explants
Gut mucosal explants from both LI and SI were processed as previously described.9 Briefly, biopsies were collected and cultured using RPMI 1640 medium with 2.5 mg/mL amphotericin B and 0.1 mg/mL piperacillin–tazobactam in a 37°C humidified incubator. Triplicate biopsies were infected with HIV-1BaL for 2 h, thoroughly washed, and replaced with fresh media on absorbable gelatin sponge in separate wells of standard tissue culture plates. Cultures were maintained for a total of 14 days. Supernatant was collected on designated days and used for p24 quantification by ELISA (SAIC-Fredrick, Inc., NCI Frederick, MA).
Statistical analyses
Phenotypic data (Figs. 1 and 2) were compared using nonparametric Wilcoxon signed-rank tests. To compare p24 replication (Fig. 4), the cumulative p24 values were log-transformed, and then, linear regression was used to determine slope of time versus log-p24 (Fig. 5). This analysis was not performed for the dose 100 TCID50 due to insufficient nonzero data points. Slopes and intercepts were compared using analysis of covariance. All statistical analyses were performed using GraphPad Prism 7 (v7.0d; GraphPad Software).
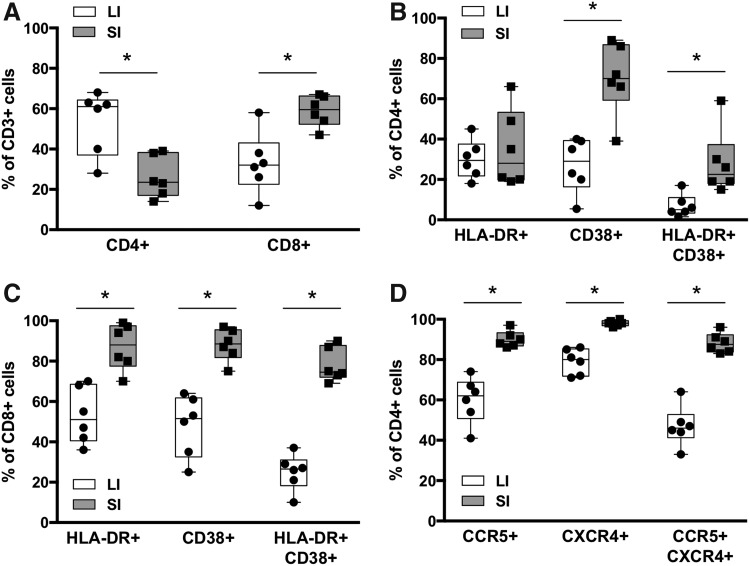
FIG. 1.
T cell phenotypes in LI and SI mucosa. (A) Percentages of CD4+ and CD8+ T cells in isolated MMCs. Activation phenotypes of (B) CD4+ T cells and (C) CD8+ T cells in isolated MMCs. (D) Expression of HIV-1 coreceptors on CD4+ T cells in isolated MMCs. The box represents the interquartile range and the whiskers extend to the minimum and maximum. The line in the box indicates the median. *p < .05 using Wilcoxon signed-rank tests. LI, large intestine; MMCs, mucosal mononuclear cells; SI, small intestine.
FIG. 2.
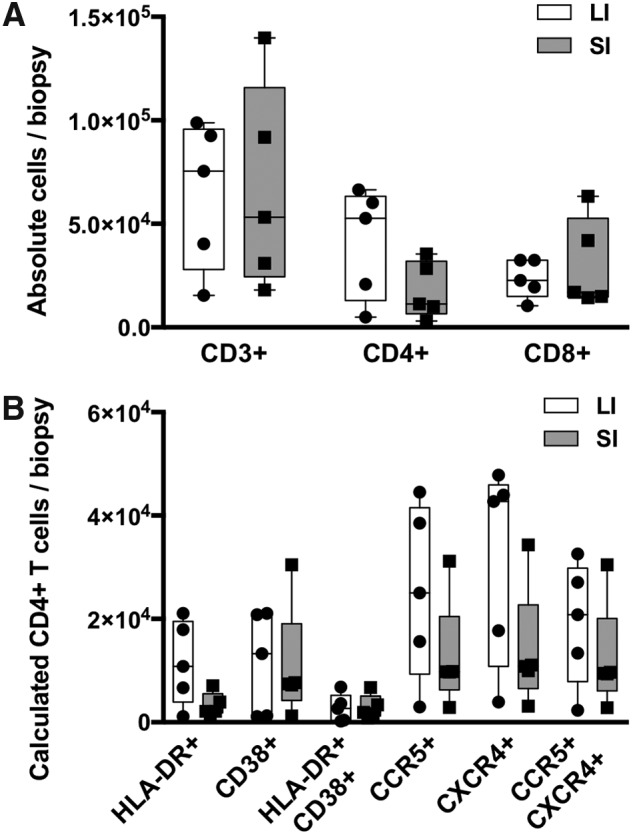
Absolute T cell counts per biopsy. Cell numbers were quantified using flow cytometry counting beads. (A) Comparative numbers of total T cells (CD3+) and CD4+ and CD8+ T cell subsets in LI and SI biopsies. (B) Comparative numbers of potential HIV-1 target cells, including activated (HLA-DR, CD38) CD4+ T cells and coreceptor (CCR5, CXCR4) expressing CD4+ T cells. The box represents the interquartile range and the whiskers extend to the minimum and maximum. The line in the box indicates the median. All differences between SI and LI biopsies were not significant using Wilcoxon signed-rank tests.
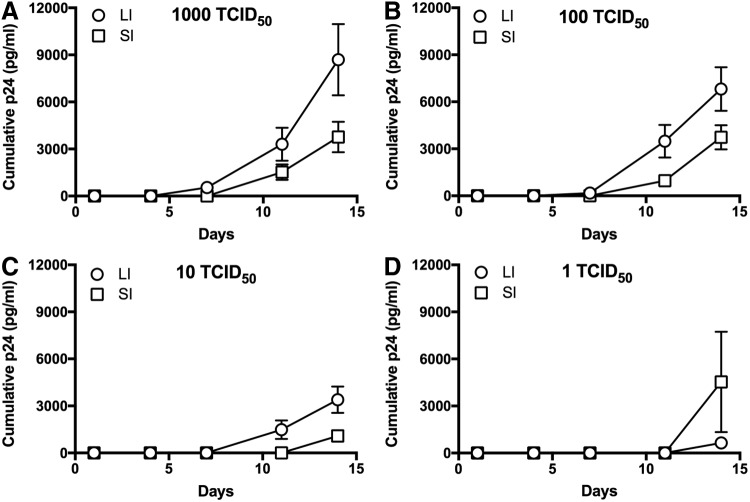
FIG. 4.
HIV-1 replication following infection with standardized doses of HIV-1. Graphs showing p24 accumulation over time since infection using (A) 103 TCID50, (B) 102 TCID50, (C) 101 TCID50, and (D) 100 TCID50 of HIV-1BaL. Data shown as mean of all infected explants per dose from all participants and error bars represent standard error.
FIG. 5.
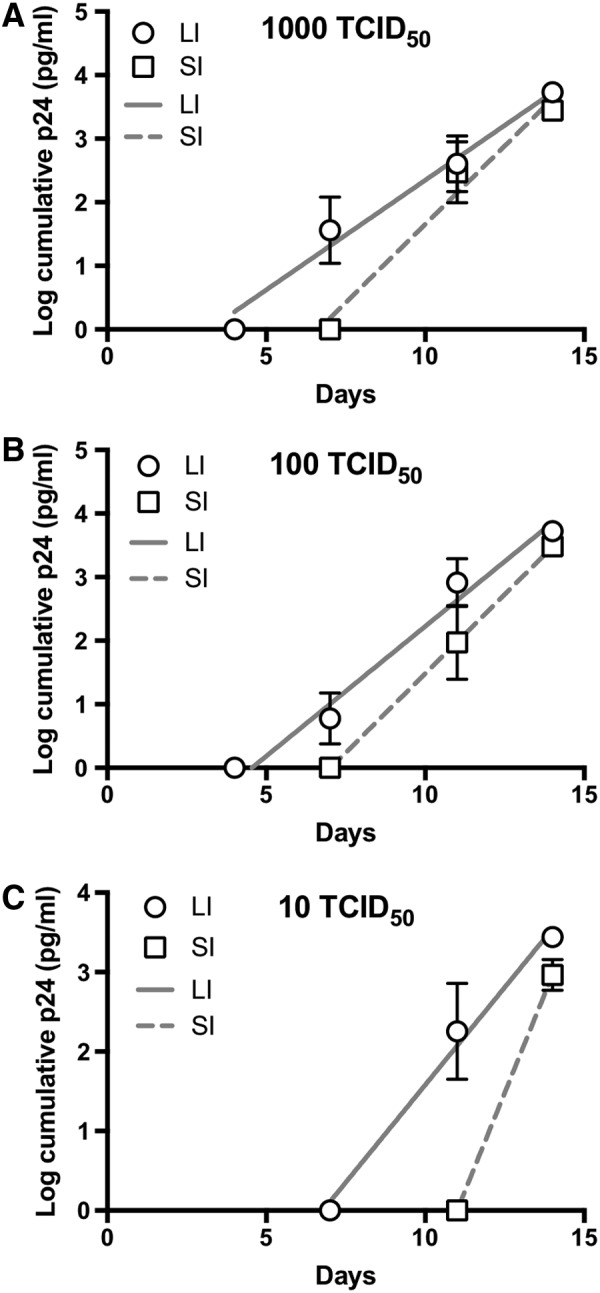
Semilog-transformed HIV-1 replication following infection with standardized doses of HIV-1. Cumulative p24 data from Figure 4 were log-transformed and then plotted versus time since infection using (A) 103 TCID50, (B) 102 TCID50, and (C) 101 TCID50 of HIV-1BaL. Lines show linear regression models for each data set (LI or SI). Semilog transformation was not performed for 100 TCID50 due to insufficient nonzero data points. Data shown as mean of all infected explants per dose from all subjects and error bars represent standard error.
Results
LI and SI have similar numbers of potential HIV-1 target cells
Biopsies were collected from HIV-seronegative participants with endoscopically normal appearing intestinal mucosa. SI samples were collected in the second portion of the duodenum (above the ampulla of Vater) and LI samples were collected between 10 and 30 cm from the anal verge. To assess the number of potential HIV-1 target cells, MMCs were isolated from both LI and SI samples and T lymphocyte quantity and phenotype were assessed by flow cytometry. The percent of total T cells (CD45+ CD3+) did not differ between LI and SI (data not shown). The percent of CD4+ T cells was greater in LI than SI (Fig. 1A), however, this difference diminished when examining the absolute number of CD4+ T cells per biopsy (Fig. 2A). Mucosal CD8+ T cells were greater in SI than LI when quantifying total percentages (Fig. 1A), but again these differences were not appreciated when the absolute number of CD8+ T cells was quantified per biopsy (Fig. 2A).
HIV-1 requires activated CD4+ T lymphocytes for efficient replication, and access to such regulates systemic dissemination.12–14 Therefore, we examined the phenotype of isolated T cells from both SI and LI compartments. The proportion of both activated CD4+ and CD8+ T lymphocytes (HLA-DR+ CD38+) was higher in the SI (Fig. 1B, C). When quantifying HIV-1 coreceptors CCR5 and/or CXCR4, nearly all (>90%) of CD4+ T lymphocytes in the SI expressed either or both coreceptors compared with only 60% in the LI (Fig. 1D). We next determined the calculated absolute cell count number for each activated subset based on the Trucount data for the CD4 populations (Fig. 2A). In doing so, we found that the number of activated CD4+ T lymphocytes was more similar between LI and SI (Fig. 2B). In addition, the calculated absolute numbers of CD4+ T lymphocytes expressing HIV-1 coreceptors CCR5 and/or CXCR4 were also similar between the compartments (Fig. 2B). This suggests that LI and SI explants contain similar numbers of potential HIV-1 target cells.
LI explants are more infectible than SI explants
Based on the phenotypic data for CD4+ T cells (Figs. 1 and 2), we hypothesized that explants from LI and SI would have similar HIV-1 infectivity and replication. Ex vivo explants were prepared using biopsies from SI and LI and then infected using limiting dilution titers (103, 102, 101, and 100 TCID50). Viral replication was quantified by the accumulation of p24 in the culture media. To assess infectivity, we used a p24 threshold value to determine infected (>100 ng) versus uninfected (<100 ng) explants.15 At the majority of titers, including clinically relevant low titers such as 101, LI explants are more easily infected as evidenced by a greater proportion of infected explants (Fig. 3A). The lowest dilution (100) data are limited by overall low infection (one LI explant vs. two SI explants).
FIG. 3.
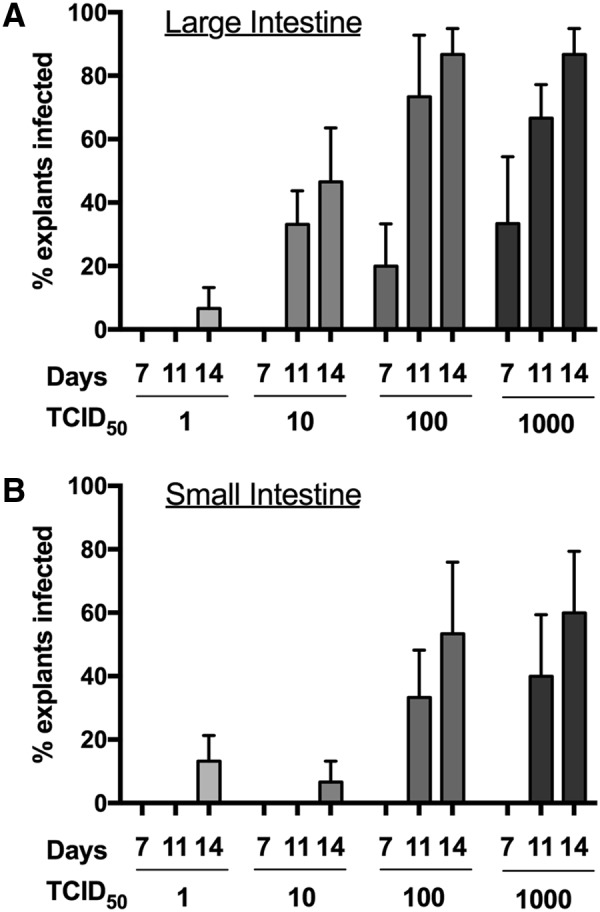
HIV-1 infection efficiency in LI and SI explants. Triplicate explants from (A) LI and (B) SI biopsies from all participants were infected with indicated doses of HIV-1BaL. Explants were considered “infected” if supernatant collected from designated day contained detectable p24 above a 100 ng/mL threshold. Data shown as the mean of the percentages of triplicate explants that met criteria for “infected” from all participants and error bars represent standard error.
LI explants exhibited earlier and greater p24 accumulation
With the greater percent infected with LI explants (Fig. 3A), we also examined the HIV-1 replication over time as this could be one potential mechanism allowing for apparent increased infectivity in these explants. As shown in Figure 4, the LI explants produced greater cumulative p24 antigen than SI explants (Fig. 4), which is a standard metric for comparative analyses of HIV-1 ex vivo infection.15–21 The LI explants consistently produced approximately threefold more p24 antigen than SI explants at the end of the measured infection period. We examined the slopes of p24 accumulation using linear regression of semilog-transformed p24 accumulation plots (Fig. 5). The slopes were not significantly different between LI and SI explants for all TCID50. However, at all TCID50 doses, the LI explants exhibited a significantly earlier rise in detectable p24 based on calculated x-intercept from the linear regression model (3.2 days vs. 6.6 days for 103, p = .05; 4.5 days vs. 7 days for 102, p = .03; 6.8 days vs. 11 days for 101, p = .006). All explants (both LI and SI) had earlier detectable p24 with higher TCID50 doses.
Discussion
In this study, we compared the HIV-1 target cell availability and ex vivo susceptibility of tissue biopsies obtained from different anatomical compartments of the human intestine: SI and the LI. We showed that although the LI had a high proportion of CD4+ T lymphocytes, the SI had greater relative proportion of activated CD4+ T cells, which are the primary target for HIV-1 infection. The absolute number of cells quantified per biopsy, however, was similar between SI and LI. When tested in ex vivo assays, the explants from the LI more efficiently established a productive infection than the SI explants.
The availability of target cells is a key determining factor during acute HIV-1 infection. Since the GI mucosa is a physiologically inflamed tissue with abundant activated CD4+ T cells expressing CCR5 and CXCR4, it is an attractive target site for virus replication. Indeed, we found that both LI and SI contain a large proportion of potential HIV-1 target cells (Figs. 1 and 2). While differences in percentages of CD4+, CD8+, and activated T cell subsets were appreciated between LI and SI samples, the absolute numbers of activated CD4+ T lymphocytes were statistically similar between the two anatomic locations. Therefore, differences in target cell number may not be sufficient to explain the observed differences in explant susceptibility and infection efficiency. Another possible consideration is the distribution of target cells. While we quantified cell numbers, we did not assess the proximity of target cells within the tissue using our methods. As cell/cell spread has been shown to be an important factor in early HIV-1 infection,22–24 it is possible that the LI had larger organized pools of activated CD4+ T lymphocytes that could sustain infection, whereas SI had more disperse populations. These findings also raise the possibility that other factors beyond target cell availability may mediate infection. One such factor could be other innate immune cells, including dendritic cells and macrophages. While these cells have been shown to facilitate HIV-1 infection of CD4+ T lymphocytes in the rectal mucosa,25,26 macrophages in the SI do not express HIV coreceptors CCR5 or CXCR4 and are therefore not permissive to HIV-1 infection.27,28 Further studies to examine these questions in relevant tissue are warranted to better understand the determinants mediating mucosal HIV-1 infection.
Our study examined the efficiency of viral infection as determined by the ability of explants to establish an infection at limiting virus concentrations, which may be a better physiologic representation. The LI explants were overall more easily infectible (Fig. 3), but of note, biopsies derived from either tissue source were ultimately capable of establishing an infection at the lowest dose of virus (100 TCID50). The physiological relevance of these findings remains to be seen, but they suggest that even the escape of a small amount of virus from the bounds of founder populations may be all that is required to establish sustained infection.
This study is subject to caveats. While flow cytometry allows for quantification of multiple simultaneous cellular subsets, it is limited in its efficiency for absolute quantification.29 Explant infections are prone to sampling error, as the location and size of the biopsy relative to a lymphoid aggregate or Peyer's patch could potentially influence infection. In an attempt to control this, all infections are performed in at least triplicate. The phenotypic studies are subject to the same error, but again biologic replicates are included to reduce this bias. A strength of our study is the within-subject comparison between the LI and SI, which reduces the influence of donor to donor variation in our comparative analysis. However, we cannot fully exclude the possibility that donor variation contributed to the observed differences in infectivity.
To our knowledge, this is the first report comparing susceptibility to HIV-1 infection in different regions of human GI mucosa. We found that both compartments contained statistically similar potential target cells, however, the LI biopsies supported more efficient HIV-1 replication. This difference is important not only for ex vivo study design but also for further investigation of the mechanisms that mediate HIV-1 mucosal infection.
Supplementary Material
Acknowledgments
The authors thank the dedicated participants in this study, Justin Akin for his assistance in preparing this article, and Terry Saunders, PA-C, for clinical assistance. They are also grateful to Dr. Robin Shattock, Dr. Leonid Margolis, and Dr. Otto Yang for helpful discussions. This work was supported, in part, by NIH/NIAID U19 AI060614 to P.A.A and I.M. The UCLA Center for AIDS Research (CFAR) Mucosal Immunology Core provided developmental funds and technical support (NIH/NIAID AI028697). J.A.F. was supported, in part, by NIH KL2 TR001882 (UCLA CTSI).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Patel P, Borkowf CB, Brooks JT, Lasry A, Lansky A, Mermin J: Estimating per-act HIV transmission risk: A systematic review. AIDS 2014;28:1509–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veazey RS, DeMaria M, Chalifoux LV, et al. : Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 1998;280:427–431 [DOI] [PubMed] [Google Scholar]
- 3.Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M: Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 2005;434:1093–1097 [DOI] [PubMed] [Google Scholar]
- 4.Li Q, Duan L, Estes JD, et al. : Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 2005;434:1148–1152 [DOI] [PubMed] [Google Scholar]
- 5.Mehandru S, Poles MA, Tenner-Racz K, et al. : Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med 2004;200:761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voigt RM, Keshavarzian A, Losurdo J, et al. : HIV-associated mucosal gene expression: Region-specific alterations. AIDS 2015;29:537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donaldson GP, Lee SM, Mazmanian SK: Gut biogeography of the bacterial microbiota. Nat Rev Micro 2016;14:20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abner SR, Guenthner PC, Guarner J, et al. : A human colorectal explant culture to evaluate topical microbicides for the prevention of HIV infection. J Infect Dis 2005;192:1545–1556 [DOI] [PubMed] [Google Scholar]
- 9.Fletcher PS, Elliott J, Grivel JC, et al. : Ex vivo culture of human colorectal tissue for the evaluation of candidate microbicides. AIDS 2006;20:1237–1245 [DOI] [PubMed] [Google Scholar]
- 10.Richardson-Harman N, Lackman-Smith C, Fletcher PS, et al. : Multisite comparison of anti-human immunodeficiency virus microbicide activity in explant assays using a novel endpoint analysis. J Clin Microbiol 2009;47:3530–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shacklett BL, Yang O, Hausner MA, et al. : Optimization of methods to assess human mucosal T-cell responses to HIV infection. J Immunol Methods 2003;279:17–31 [DOI] [PubMed] [Google Scholar]
- 12.Miller CJ, Li Q, Abel K, et al. : Propagation and dissemination of infection after vaginal transmission of simian immunodeficiency virus. J Virol 2005;79:9217–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Schuler T, Zupancic M, et al. : Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 1999;286:1353–1357 [DOI] [PubMed] [Google Scholar]
- 14.Zhang ZQ, Wietgrefe SW, Li Q, et al. : Roles of substrate availability and infection of resting and activated CD4+ T cells in transmission and acute simian immunodeficiency virus infection. Proc Natl Acad Sci U S A 2004;101:5640–5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson-Harman N, Mauck C, McGowan I, Anton P: Dose-response relationship between tissue concentrations of UC781 and explant infectibility with HIV type 1 in the RMP-01 rectal safety study. AIDS Res Hum Retroviruses 2012;28:1422–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson-Harman N, Hendrix CW, Bumpus NN, et al. : Correlation between compartmental tenofovir concentrations and an ex vivo rectal biopsy model of tissue infectibility in the RMP-02/MTN-006 phase 1 study. PLoS One 2014;9:e111507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anton PA, Saunders T, Elliott J, et al. : First phase 1 double-blind, placebo-controlled, randomized rectal microbicide trial using UC781 gel with a novel index of ex vivo efficacy. PLoS One 2011;6:e23243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bunge AKE, Dezzutti CS, Rohan LC, et al. : A Phase 1 trial to assess the safety, acceptability, pharmacokinetics and pharmacodynamics of a novel dapivirine vaginal film. J Acquir Immune Defic Syndr (1999) 2016;71:498–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dezzutti CS, Russo J, Wang L, et al. : Development of HIV-1 rectal-specific microbicides and colonic tissue evaluation. PLoS One 2014;9:e102585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrera C, Cranage M, McGowan I, Anton P, Shattock RJ: Colorectal microbicide design: Triple combinations of reverse transcriptase inhibitors are optimal against HIV-1 in tissue explants. AIDS 2011;25:1971–1979 [DOI] [PubMed] [Google Scholar]
- 21.Scott YM, Park SY, Dezzutti CS: Broadly neutralizing anti-HIV antibodies prevent HIV infection of mucosal tissue ex vivo. Antimicrob Agents Chemother 2016;60:904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boullé M, Müller TG, Dähling S, et al. : HIV cell-to-cell spread results in earlier onset of viral gene expression by multiple infections per cell. PLoS Pathog 2016;12:e1005964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwami S, Takeuchi JS, Nakaoka S, et al. : Cell-to-cell infection by HIV contributes over half of virus infection. Elife 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sigal A, Kim JT, Balazs AB, et al. : Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature 2011;477:95–98 [DOI] [PubMed] [Google Scholar]
- 25.Cameron PU, Freudenthal PS, Barker JM, Gezelter S, Inaba K, Steinman RM: Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science 1992;257:383–387 [DOI] [PubMed] [Google Scholar]
- 26.Gurney KB, Elliott J, Nassanian H, et al. : Binding and transfer of human immunodeficiency virus by DC-SIGN+ cells in human rectal mucosa. J Virol 2005;79:5762–5773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng G, Sellers MT, Mosteller-Barnum M, Rogers TS, Shaw GM, Smith PD: Lamina propria lymphocytes, not macrophages, express CCR5 and CXCR4 and are the likely target cell for human immunodeficiency virus type 1 in the intestinal mucosa. J Infect Dis 2000;182:785–791 [DOI] [PubMed] [Google Scholar]
- 28.Li L, Meng G, Graham MF, Shaw GM, Smith PD: Intestinal macrophages display reduced permissiveness to human immunodeficiency virus 1 and decreased surface CCR5. Gastroenterology 1999;116:1043–1053 [DOI] [PubMed] [Google Scholar]
- 29.Preza GC, Yang O, Elliott J, Anton P, Ochoa MT: T lymphocyte density and distribution in human colorectal mucosa, and inefficiency of current cell isolation protocols. PLoS One 2015;10:e0122723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.