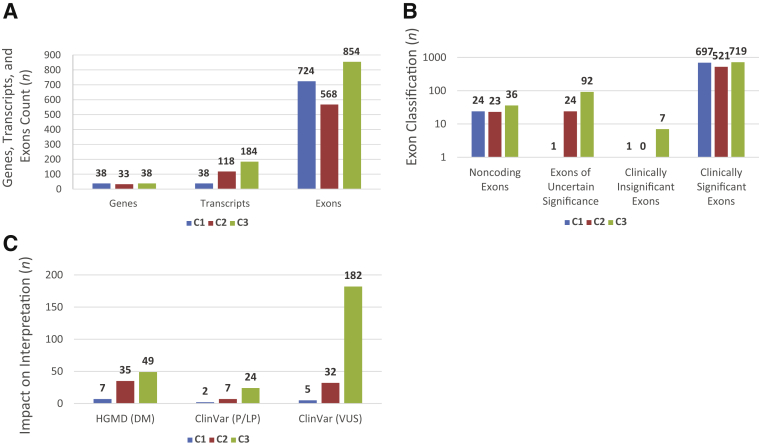
Figure 2.
A: Gene, exon, and transcript counts: genes were categorized [category 1 (C1), category 2 (C2), and category 3 (C3)], and transcripts and exons were counted. B: Exon counts across categories: for each of the three categories, exons were classified as Clinically significant, uncertain significance, clinically insignificant, or noncoding, as per the definitions in Figure 1C. C: Impact on interpretation: variant counts in uncertain and insignificant exons for each category were collected from the Human Gene Mutation Database (HGMD) and ClinVar. DM, disease-causing mutation; LP, likely pathogenic; P, pathogenic; VUS, variant of uncertain significance.