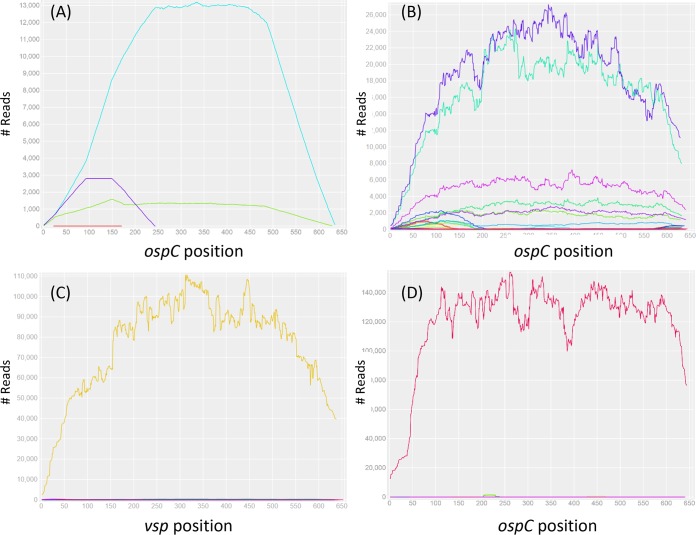
FIG 2.
Read depths of ospC alleles in simulated and tick samples.(A) Simulated reads were generated on the basis of nucleotide sequences of two ospC alleles (F and J) which were subsequently mixed in a 1F:10J proportion. The reads were aligned to all 20 reference sequences (see Section S2 in the supplemental material), and only the two input alleles (J, cyan; F, green; E, red; K, purple) showed complete full-length coverages and approximately the same input proportions, validating the specificity and sensitivity of the bioinformatics protocol for allele identification (a full test of specificity is shown in Section S4). (B) Seven ospC alleles (O, I, U, H, T, E, and K) were detected in an adult tick (M119, male, Rockefeller State Park [RF], fall of 2016). (C) The universal ospC primer set was able to amplify not only the ospC locus in Borreliella species but also the vsp locus in Borrelia miyamotoi (see alignment in Section S1). Here, the vsp locus was detected in a nymph tick (N030, RF, summer of 2015). (D) A previously unknown ospC allele (C14, GenBank accession MH071431) was detected in a nymph tick (N150, RF, summer of 2016), suggesting the presence of a new B. bissettiae-like species.