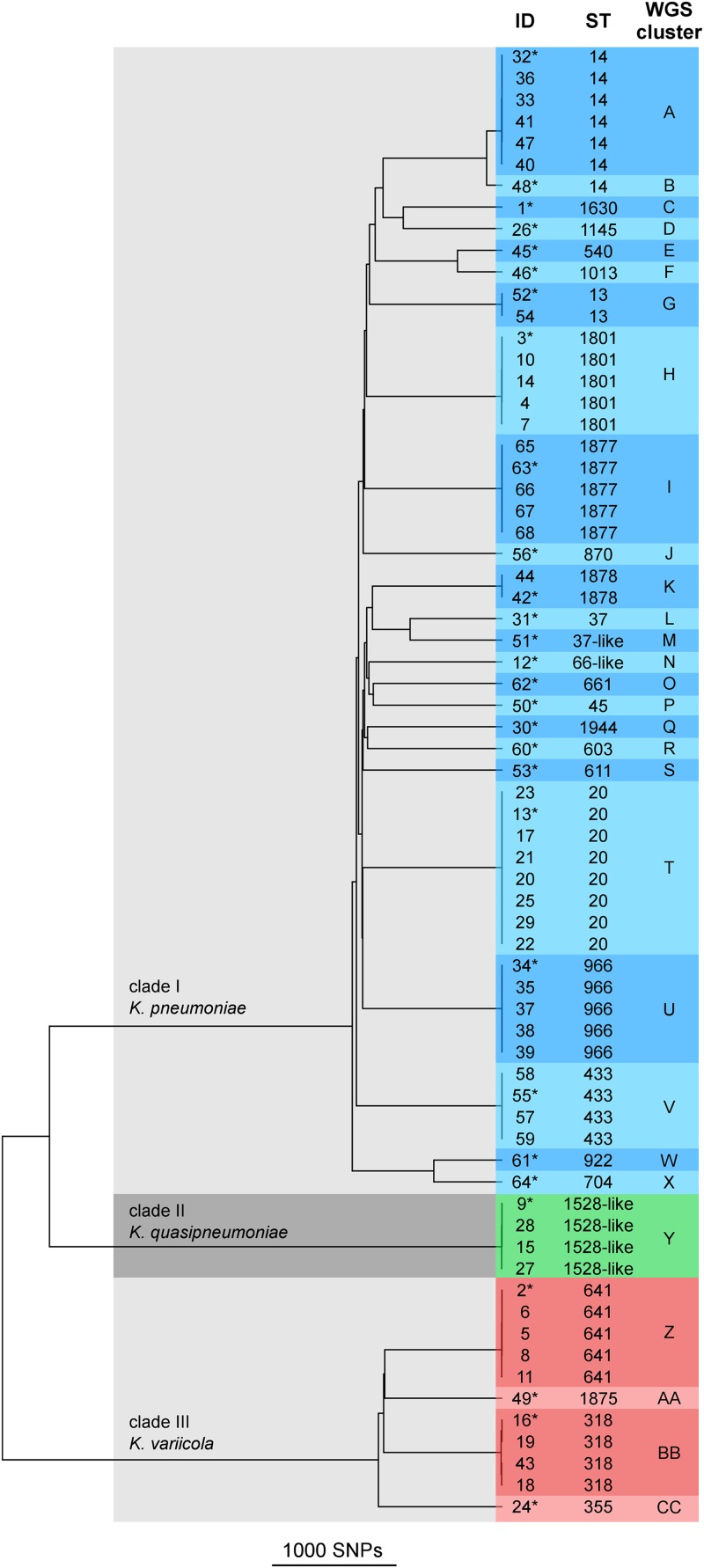
FIG 1.
SNP-based cluster dendrogram of 68 Klebsiella isolates. A core genome of all genome sequences was used to call SNPs in the isolate genomes. Genomes with distances of <18 SNPs were assigned to the same WGS cluster (colored boxes). The multilocus sequence type (ST) was extracted from the assembled genome sequences. Clade assignment and species identification were derived from average nucleotide identity analysis (Table S2 in the supplemental material) of one representative isolate (indicated by an asterisk) from each cluster.