Plasmodium falciparum malaria pathogenesis is tied to the sequestration of parasites in the microvasculature. Parasite sequestration leading to severe malaria is mediated by P. falciparum erythrocyte membrane protein 1 (PfEMP1) binding to endothelial protein C receptor (EPCR) via its CIDRα1 domains.
KEYWORDS: EPCR, antigen diversity, CIDRα1, cross-reactive antibody, malaria, PfEMP1, vaccine
ABSTRACT
Plasmodium falciparum malaria pathogenesis is tied to the sequestration of parasites in the microvasculature. Parasite sequestration leading to severe malaria is mediated by P. falciparum erythrocyte membrane protein 1 (PfEMP1) binding to endothelial protein C receptor (EPCR) via its CIDRα1 domains. CIDRα1 domains are targets of naturally acquired immunity, and a vaccine eliciting antibodies inhibiting the EPCR binding of CIDRα1 could potentially prevent disease and death from malaria. CIDRα1 domains have diversified in sequence to escape immune recognition but preserved structure to maintain EPCR binding. The EPCR-binding CIDRα1 domains separate into six major sequence types predicted to form a conserved structure in which only the amino acids essential for EPCR binding are highly conserved. Here, we investigated whether antibodies elicited by vaccination with single or multiple recombinant CIDRα1 domains are able to bind and inhibit diverse CIDRα1 domains. We found that EPCR binding-inhibitory antibodies to CIDRα1 variants closely related to those used for vaccination are readily elicited, whereas antibodies binding distant CIDRα1 variants are sporadically generated and are rarely inhibitory. Despite this, sequence similarity correlated poorly with the ability of induced antibodies to inhibit across diverse variants, and no continuous sequence regions of importance for cross-inhibitory antibodies could be identified. This suggested that epitopes of cross-variant inhibitory antibodies were predominantly conformational. Vaccination with immunogens engineered to focus immune responses to specific epitopes or an optimal choice of multiple CIDRα1 variants may improve elicitation of broadly reactive and inhibitory antibody responses.
INTRODUCTION
The clinical outcome of Plasmodium falciparum infections is linked to the sequestration of infected erythrocytes in the host microvasculature (1). Parasites export the P. falciparum erythrocyte membrane protein 1 (PfEMP1) adhesion molecules to the surface of infected erythrocytes, where they bind specific human endothelial receptors and facilitate withdrawal of the parasites from blood circulation and escape from splenic clearance (2). In addition to ensuring parasite survival and allowing exponential growth of the blood-stage parasites, the accumulation of parasites in host organs contributes to pathogenesis by occluding blood flow and inducing a strong and, to the host, sometimes harmful inflammatory response (3–5). Together, these processes can lead to multiple and often overlapping symptoms, including severe anemia, respiratory distress, and neurological impairment (cerebral malaria), all included in the collective term severe malaria (6, 7).
Immunity to malaria develops as a result of repeated infections, with immunity to severe malaria developing first (8, 9). In areas with high transmission of P. falciparum, immunity to severe malaria is acquired early in life, whereas immunity to uncomplicated malaria outcomes is developed later, and protection from infection is never achieved. A large body of immunoepidemiological evidence (recently reviewed in Bull and Abdi [10]) has converged on a subset of PfEMP1 molecules being the main target of IgG protecting against severe malaria. Recent studies of PfEMP1 gene expression in patients have identified the subset of PfEMP1 molecules associated with development of severe malaria symptoms, including severe anemia and cerebral malaria (11–21). This subset is defined by carrying a so-called CIDRα1 domain mediating tethering to endothelial cells through binding to endothelial protein C receptor (EPCR) (22). These observations make CIDRα1 domains a main target to be included in vaccines aiming to elicit immune responses conferring protection against malaria.
However, the antigenic diversity of CIDRα1 poses a significant challenge for the development of such a vaccine. Through evolution, CIDRα1 molecules have diversified in sequence to escape immune recognition but have retained their overall structure for high-affinity binding to EPCR (23). Intriguingly, the molecular mechanism of the CIDRα1-EPCR interaction closely mimics the mechanism of the interaction between EPCR and its natural ligand, activated protein C. This and the fact that the interaction is ancient (also found in Plasmodium reichenowi parasites infecting chimpanzees) (24) suggest that the host-parasite interaction has reached an evolutionary state in which CIDRα1 molecules can vary in sequence but not overall structure without also reducing affinity to EPCR and, ultimately, parasite survival. CIDRα1 domains cluster by sequence similarity into subtypes, named CIDRα1.1 to CIDRα1.8 (Fig. 1), all of which, with a few exceptions, bind EPCR with high affinity. These exceptions are CIDRα1.5b domains, which exhibit a distinct sequence deviation across the EPCR binding site, and the two minor groups CIDRα1.2 and -1.3, found in the var1 pseudogenes. Distinct subsets of PfEMP1-encoding genes are maintained by a recombination hierarchy imposed by a chromosomal organization of the genes (25). The closely related CIDRα1.1 and CIDRα1.8 domain subtypes are found in a subset of genes known as group B/A or cassette 8 (DC8) genes, due to their unique domain composition. The domain subtypes CIDRα1.4 to CIDRα1.7 are encoded by so-called group A genes (23, 24). Diversity within each CIDRα1 domain subtype is significant, but studies of IgG from malaria-exposed individuals indicate that genuinely cross-reactive and broadly inhibitory antibodies are developed in response to infection (23, 26). In this study, we explored the immunogenicity of CIDRα1 domains by immunizing animals with different recombinant CIDRα1 domains and testing the reactivity and EPCR binding-inhibitory effect of the elicited antibodies on a panel of recombinant CIDRα1 domains.
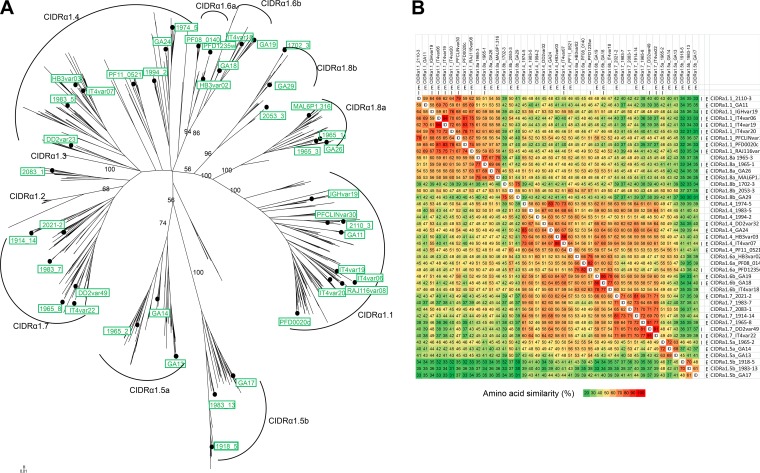
FIG 1.
Sequence similarity of PfEMP1 CIDRα1 domains. (A) Maximum likelihood tree (key bootstrap [n = 50] values are indicated on branches) of 885 CIDRα1 sequences (30 kDa) (generated in Lau et al. [23]) with the 43 variants included in this study either as immunogen or in antibody. Reactivity measurements are marked by dots, and variant names are given in green boxes. Named arches mark CIDRα1 sequence subtypes as defined in Lau et al. (23). (B) Pairwise amino acid sequence identity (ID, identical sequence) heat map of the CIDRα1 recombinant domains (19 kDa) used in this study as immunogens (I) and for antibody detection in ELISA (E) or in Luminex (all proteins listed) assays.
RESULTS
Immunization with diverse single EPCR-binding recombinant CIDRα1 domains.
Groups of four rats were immunized with four different 19-kDa Strep-tag II (STRPII)-tagged recombinant CIDRα1 domains. After three immunizations 3 weeks apart, plasma was collected, and IgG reactivity to 43 30-kDa His-tagged recombinant CIDRα1 domain variants representing all CIDRα1 sequence subgroups (Fig. 1) was measured using a bead-based (Luminex) multiplex assay (Fig. 2). IgG reacting with the CIDRα1 variants used as immunogens was elicited in animals. The CIDRα1 variants used for immunization also induced IgG against other CIDRα1 domains but to different degrees and most effectively to domains within the same subtype as that of the immunogen. This was particularly clear for the CIDRα1.1 domain, where immunization with CIDRα1.1_IT4var20 induced high levels of IgG reacting with other CIDRα1.1 subtypes as well as their closest related variants, the CIDRα1.8 domains. Most animals immunized with CIDRα1.1_IT4var20 also had IgG-reacting domain variants CIDRα1.4 to CIDRα1.7. Regardless of the CIDRα1 subtype of the immunogen, induction of IgG reactive with domains binding CD36 was erratic. These data indicated that vaccination with a single CIDRα1 domain elicited IgG that was reactive across several domains. We next tested if the cross-reactive IgG was functional in inhibiting binding between CIDRα1 and EPCR.
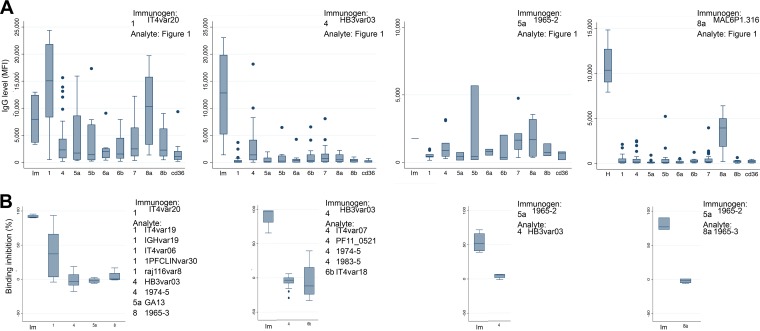
FIG 2.
CIDRα1 reactivity and EPCR binding inhibition of IgG from animals immunized with a single recombinant CIDRα1 domain. The subtypes and specific variants of the CIDRα1 domains used as immunogens and analytes are stated in each panel (“Analyte: Figure 1” indicates that IgG reactivity was measured against the 43 recombinant CIDRα1 domains listed in Fig. 1B). (A) The IgG reactivity to the 43 CIDRα1 domains (as mean fluorescent intensity [MFI] measured by Luminex assay) is shown grouped by the domain used for immunization (Im) and by the subtype of the analytes: 1, CIDRα1.1; 4, CIDRα1.4; 5a or 5b, CIDRα1.5; 6a or 6b, CIDRα1.6; 7, CIDRα1.7; 8a or 8b, CIDRα1.8; cd36, CD36-binding CIDR. Box plots show median reactivity with 25th and 75th percentiles, upper and lower adjacent values, and outliers of plasma from four immunized animals to each of the tested domains. (B) The abilities of elicited IgGs to inhibit EPCR binding (ELISA) of the domain used for immunization (Im) and other select CIDRα1 domains are shown grouped by the CIDRα1 subtype of the analytes, as indicated on the x axes.
For these assays we purified IgG from immunized animals before testing the ability of the IgG to inhibit binding between EPCR and 30-kDa CIDRα1 domains (Fig. 2). IgG inhibiting EPCR binding of the CIDRα1 variant used for immunization was readily induced. However, the induction of cross-inhibitory IgG was sporadic and mainly found in animals immunized with the CIDRα1.1_IT4var20 domain and in which the EPCR binding of other CIDRα1.1 domains was inhibited.
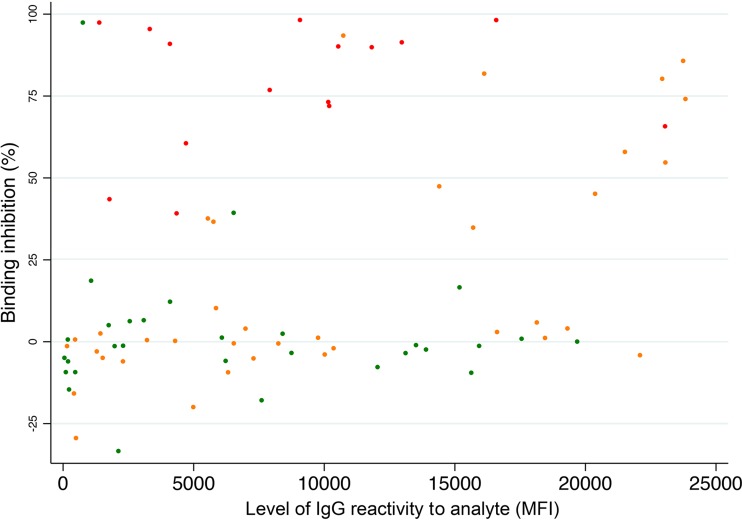
We then investigated if overall IgG reactivity against a domain predicted the ability of the IgG to inhibit EPCR binding of the domain (Fig. 3) and found a weak positive correlation (Rs = 0.35, P = 0.002, Spearman's rank correlation). However, the relationship was by no means absolute, and a high level of inhibitory activity was sometimes measured in plasma with a relatively low level of IgG recognition. This indicated that the inhibitory IgG constituted a minor fraction of the induced IgG.
FIG 3.
Relation between plasma reactivity with a CIDRα1 domain (determined as mean fluorescent intensity [MFI]) measured by Luminex assay) and the ability of IgG purified from the plasma to inhibit EPCR binding of that domain. Red, immunogen and analyte variants are identical; orange, immunogen and analyte variant belong to same CIDRα1 subtype; green, immunogen and analyte variant are of different CIDRα1 subtypes.
Immunization with cocktails of recombinant CIDRα1 domains of different subgroups.
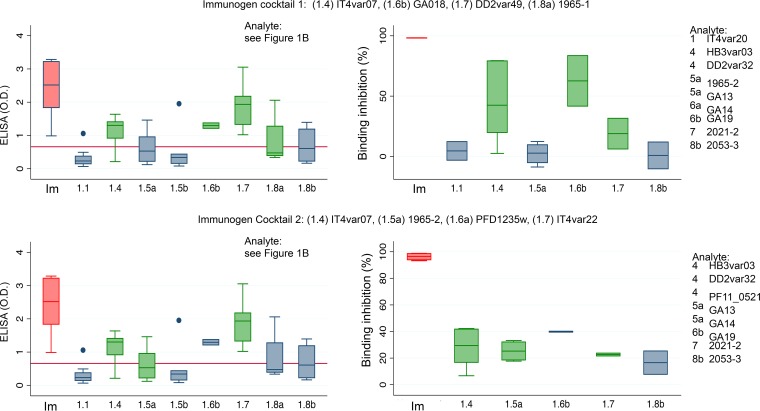
To investigate if immunization with a cocktail of different CIDRα1 variants would broaden the anti-CIDRα1 subtype responses, groups of four rats were immunized with two different cocktails containing four 30-kDa CIDRα1 domains of different subtypes (Fig. 4). This resulted in a robust induction of IgG toward the proteins present in the immunogen, in a modest induction of IgG toward proteins not included as immunogens but of the same CIDRα1 subtype, and in poor induction of IgG reactivity with CIDRα1 domains of subtypes not represented by the immunogens. Similarly, a clear EPCR binding-inhibitory effect was seen for domains included as immunogens, at best a very modest inhibition was seen of domains of the same subtype as the immunogen, and a very limited binding-inhibitory effect was seen on CIDRα1 subtypes not represented by the immunogen (Fig. 4).
FIG 4.
Antibody reactivity and EPCR binding inhibition of IgG from animals immunized with cocktails of four different CIDRα1 domains as indicated. IgG reactivity to 31 different CIDRα1 domains (analytes marked E” in Fig. 1B) was measured by ELISA (as optical density [OD]). Analytes used for testing EPCR binding inhibition are listed with subtypes and specific sequence variants. Data are shown grouped by the domain used for immunization (Im) and by the CIDRα1 subtype of the analyte, e.g., CIDRα1.1 (1.1). Data for immunogens are shown in red; data for which the immunogen and analyte CIDRα1 are overlapping or not overlapping are shown in green or blue, respectively. Box plots show median reactivity with 25th and 75th percentiles and upper and lower adjacent values. For these assays His-tagged CIDRα1 domains were used both for immunization and for the ELISAs, and plasma was depleted for His-IgG prior to ELISAs. Red lines on the left panels indicate the levels of residual anti-His-IgG in the plasma.
Immunization with cocktails of recombinant CIDRα1 domains of the same subtype.
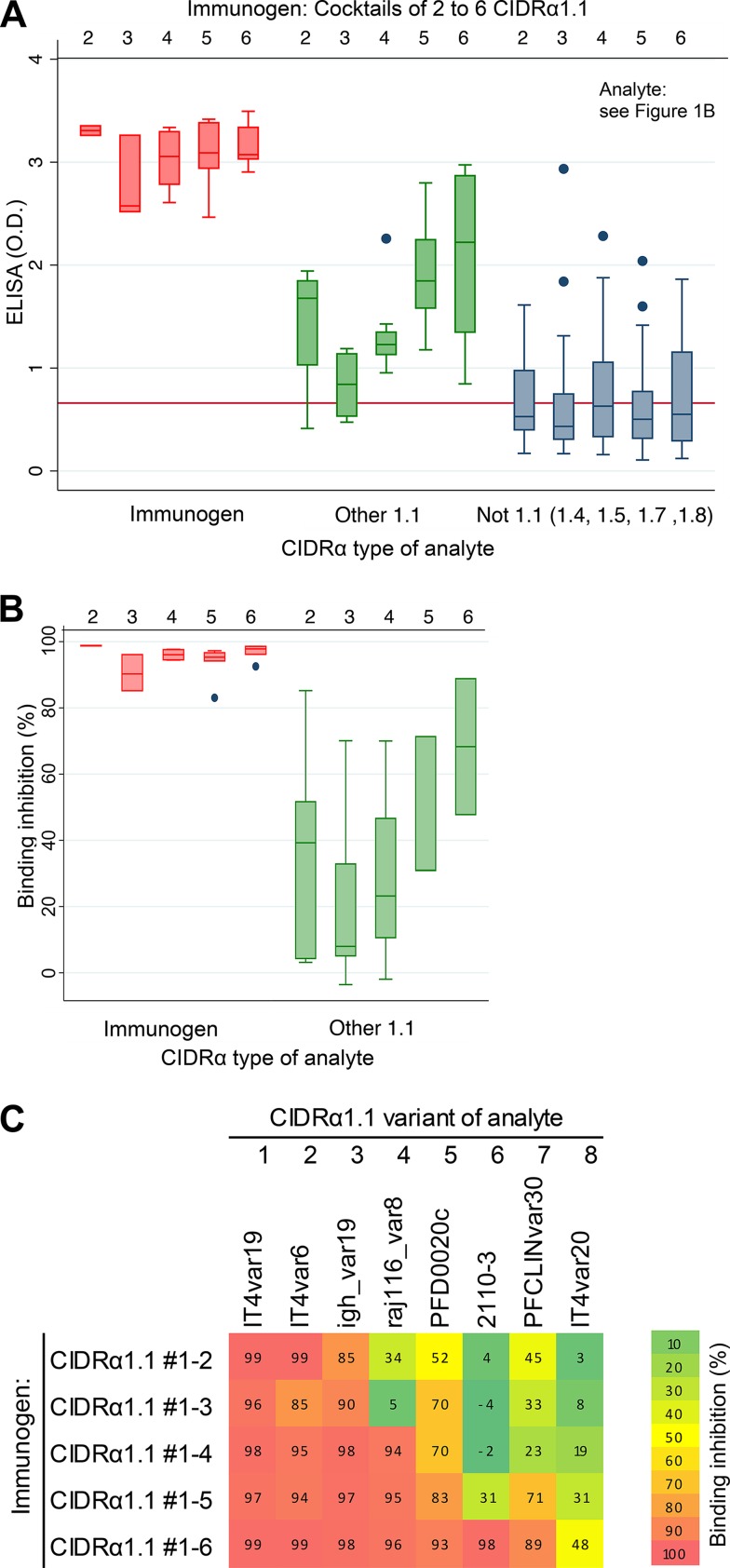
The result obtained by immunization with single domains or with cocktails of domains indicated that it was difficult to induce IgG inhibiting the interaction between EPCR and CIDRα1 domains not present in the immunogen. However, immunization with CIDRα1.1 domains did elicit some cross-inhibitory IgG (Fig. 2B, left panel). CIDRα1.1 variants represent a large and distinctly separate subtype of CIDRα1 domains. We next investigated the IgG response from animals (two rats per group) immunized with cocktails of two, three, four, five, or six CIDRα1 domains (30 kDa) (Fig. 5).
FIG 5.
Reactivity (A) and EPCR binding inhibition (B) of IgG from animals immunized with protein cocktails containing an increasing number of CIDRα1.1 domains, as indicated. Two rats were immunized per antigen cocktail containing between two and six CIDRα1.1 domains (upper horizontal numbering). Antibody reactivity was measured against 31 recombinant CIDRα1 domains (marked E in Fig. 1B): CIDRα1.1 domains present in the immunogen cocktail (immunogen), CIDRα1.1 domains not present on the cocktail (other 1.1), and CIDRα1 domains of subtypes 1.4 to 1.8 (not 1.1). IgG level was measured by ELISA (as optical density [OD]). Box plots show median reactivity with 25th and 75th percentiles and upper and lower adjacent values. For these assays, His-tagged CIDRα1 domains were used both for immunization and for the ELISAs, and plasma was depleted for His-IgG prior to ELISAs. Red lines on left panels indicate levels of residual anti-His-IgG in the plasma. (C) Association between the ability of plasma to inhibit EPCR binding of a given CIDRα1 domain (percent binding inhibition, according to the color map) and the number of CIDRα1.1 domains present in the immunogen cocktail. The eight recombinant CIDRα1.1 domains used for binding inhibition experiments are listed above the heat map, and the composition of immunogen cocktails containing between two and six of these CIDRα1.1 domains is indicated to the left of the panel. The heat map indicates that the rats immunized with six CIDRα1 domains elicited binding-inhibitory IgG against domains 7 and 8 not present in the immunogen.
IgG responses to the domains present in the immunization cocktail were robust and appeared unaffected by the coadministration of several CIDRα1.1 domains. The induction of IgG against CIDRα1.1 domains not included in the immunizations increased when additional CIDRα1.1 domains were included in the immunization cocktail. In contrast, the level of IgG reactive with types CIDRα1.4 to CIDRα1.8 was low and did not increase when additional CIDRα1.1 domains were included. Evaluation of EPCR binding inhibition of purified IgG (Fig. 5B and C) showed strong inhibition of the domains included in the immunization cocktail regardless of the number of variants included in the cocktail. There was also an increase in inhibition of EPCR binding by CIDRα1.1 domains not present in the immunization cocktail when the number of domains in the cocktail increased, albeit this increase appeared to depend on addition of specific variants in the immunogen cocktail. For example, while there was an incremental increase in inhibitory effect of CIDRα1.1_IT4var20 with addition of each extra variant in the immunogen cocktail, inhibition of igh_var19, PFD0020c, and PFCLINvar30 CIDRα1.1 variants appeared to depend on inclusion of specific variants in the immunogen cocktail. However, no sequence relation could explain these observations. These results suggest that immunization with a finite number of CIDRα1.1 domains may elicit IgG inhibiting EPCR binding of all CIDRα1.1 variants.
Immunization with single CIDRα1.1 domains.
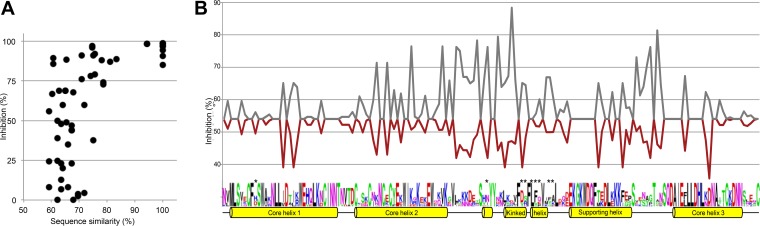
To further investigate the relation between sequence similarity and elicitation of cross-inhibitory IgG, groups of two animals were immunized with seven different CIDRα1.1 domains (all CIDRα1.1; as listed in Fig. 5C, except for the PFD0020c variant), and the ability of the elicited IgG to inhibit EPCR binding of each of eight CIDRα1.1 variants was tested. The observed level of inhibition of each of these experiments (56 data points) is plotted in Fig. 6 in relation to the sequence similarity between the domain used as immunogen and the domain inhibited. There was no clear association between sequence similarity and binding inhibition, but when sequence similarity was above 80%, inhibition was >75% (Fig. 6A).
FIG 6.
Inhibition of EPCR binding by IgG elicited after immunization with single CIDRα1.1 domains. Inhibition of binding of eight different CIDRα1.1 domains to EPCR was tested using seven separate pools of IgG purified from groups of two animals immunized with one of seven different CIDRα1.1 domains. (A) Relation between EPCR binding inhibition and the pairwise amino acid sequence identity between the immunogen and inhibited CIDRα1.1 variant. (B) Mean EPCR binding inhibition of eight CIDRα1.1 domains by the seven pools of IgG at each amino acid position in the CIDRα1.1 domain. For each amino acid position in the CIDRα1.1 sequence, the mean of the inhibition data from all immunogen and analyte pairs having either the same (gray) or a different (red) amino acid is shown. The sequence logo represents the diversity of the eight included CIDRα1.1 variants. *, amino acids predicted to directly interact with EPCR. Alpha helices and their function in the CIDRα1 domain (23) are shown.
Next, the inhibitory effect was related to the amino acid similarity at each position in the CIDRα1.1 sequence. From sequence alignments, each amino acid position was identified as either identical or different between each pair of immunogen and inhibited CIDRα1.1 variant. For each position the mean level of inhibition between pairs of domains with identical amino acids and the mean level of inhibition between pairs of domains with different amino acids were calculated. Thus, at positions with 100% conserved residues, the mean inhibition was the average of all 56 data points. These inhibition data were plotted along an annotation of the structural characteristics of CIDRα1 domains (Fig. 6B). Interestingly, the curves for the two graphs largely mirrored each other. The two curves separated most widely in the regions adjacent to the EPCR binding region and to a lesser degree at the amino acid positions involved in EPCR binding. This indicated that sequence variation across the whole domain influenced the protein interaction-inhibitory effects seen, although the highly diverse loop regions on each side of the interacting kinked alpha helix may have contributed most to the variation in inhibition.
DISCUSSION
The CIDRα1-mediated attachment of infected erythrocytes to EPCR is a potential target for vaccines to prevent severe malaria. Characterization of PfEMP1 expressed in severe malaria indicates that all EPCR-binding CIDRα1 subtypes can precipitate disease (11–21). Thus, an effective malaria vaccine targeting PfEMP1 probably has to target most, if not all, CIDRα1 subtypes. Their sequence diversity is a major challenge for vaccine development. The study presented here was conducted to assess to what degree experimental vaccination with CIDRα1 proteins elicits IgG inhibiting EPCR binding of CIDRα1 types not present in the immunogen. To enable this assessment, the study relied on the interaction between recombinant CIDRα1 proteins and EPCR for quantification of binding inhibition. This approach was chosen as binding inhibition experiments employing a large set of parasite lines expressing different CIDRα1 subtypes would be logistically challenging. We have previously demonstrated that immunization with recombinant CIDRα1 domains elicits antibodies reacting with cognate proteins natively expressed on the surface of parasitized erythrocytes and inhibits their interaction with EPCR (22).
PfEMP1 domains binding EPCR separate into distinct sequence subgroups, CIDRα1.1 and CIDRα1.4 to CIDRα1.8. There is considerable sequence variation within these groups (24). However, all sequences of EPCR-binding CIDRα1 domains align tightly around conserved amino acids, predicted to fold the proteins to present a short, ∼25-amino-acid-long, broken and kinked alpha helix structure that protrudes from a core triple helix structure (23). This 19-kDa subdomain of CIDRα1 binds EPCR mainly through amino acids present in the short kinked helix. Across the 19-kDa region, CIDRα1 sequences are pairwise from 40 to 100% identical. The average sequence identity within CIDRα1 subtypes (e.g., CIDRα1.1) is 69 to 72%, but short amino acid stretches of high similarity are shared between members of different subtypes.
Previous studies have shown that individuals in areas of malaria transmission develop cross-reactive anti-CIDRα1 antibodies, including IgG, that effectively inhibit EPCR binding. Such CIDRα1 antibodies are acquired early in life during the first couple of P. falciparum infections (23, 26). It is possible that monoclonal anti-CIDRα1 antibodies can be divided into those that can inhibit EPCR binding by binding epitopes at or near the EPCR binding site (named functional IgG here) and those that bind other epitopes and cannot inhibit EPCR binding. The latter could opsonize parasites for phagocytosis (27), but the ideal vaccine would induce IgG that inhibits EPCR binding of most CIDRα1 variants. This could be achieved by induction of a polyvariant response analogous to what is achieved by the vaccines against Streptococcus pneumoniae. However, it is also possible that some IgGs can recognize the common shape of CIDRα1 responsible for EPCR binding and thereby bind and inhibit EPCR binding of all CIDRα1 domains in a similar fashion as the conserved EPCR molecule that interacts with the diverse CIDRα1 domains. A CIDRα1 vaccine could therefore aim to induce either a broadly inhibitory IgG or a polyvalent IgG response functional across most CIDRα1 variants.
In this study, we explored to what extent cross-reactive and inhibitory antibodies were elicited by vaccination with recombinant CIDRα1 proteins. Initially, animals were immunized with a single CIDRα1 domain. This elicited antibodies with high reactivity and an inhibitory effect on the variant used as immunogen. Reactivity to other variants was mainly observed toward variants of the same CIDRα1 subtype as the immunogen, albeit high reactivity to more distant variants was seen sporadically. However, cross-reactivity was poorly associated with inhibition, and cross-inhibition was mainly observed for analytes with the same CIDRα1 subtype as the immunogen. When animals were immunized simultaneously with four CIDRα1 domains of different subgroups, the pattern was identical. The elicited IgG reacted and strongly inhibited EPCR binding of domains present in the immunogen whereas recognition and inhibition of other variants were disappointing. These results indicated that broadly inhibitory antibodies are not readily induced and that immunization can induce IgGs recognizing epitopes shared by distantly related CIDRα1 subtypes although these antibodies most often do not inhibit EPCR binding. This could be because epitopes recognized by inhibitory antibodies are less accessible or less immunogenic than the epitopes targeted by the nonfunctional antibodies. Another possibility is that the binding of inhibitory IgG molecules is highly specific and targets amino acids with structures and chemical characteristics that differ between CIDRα1 variants. With the immunization protocol employed, cross-inhibitory responses were detected mainly between CIDRα1 sequence variants with high sequence similarity.
To explore this further, the next set of experiments studied cross-reactivity within the subtype of CIDRα1.1 domains. Immunization with an increasing number of CIDRα1.1 variants increased the degree of EPCR binding inhibition of CIDRα1.1 variants not included in the immunization. As expected, potent EPCR binding inhibition was observed when there was high sequence similarity (<80%) between a domain used for immunization and the domain tested for EPCR binding. However, EPCR binding inhibition was occasionally also observed with sequence identities between 60 and 80%. Thus, in some instances there was a high degree of EPCR binding inhibition despite relatively low overall sequence similarity, but it was not possible to identify particular domain regions where a high similarity over a particular stretch of amino acids could explain the cross-inhibition observed. These results indicate that inhibitory antibodies targeting the immunogens are readily elicited and that epitopes recognized by cross-variant inhibitory antibodies, while dependent on amino acid similarity, predominantly are conformational. Further characterization of the epitopes recognized by functional and nonfunctional monoclonal antibodies may aid design of engineered immunogens eliciting a broadly reactive functional IgG, similar to what has been achieved for engineered versions of the closely related Plasmodium vivax Duffy binding proteins (28).
These data suggest that broadly reactive and inhibitory antibodies are not readily induced or at least are not a prominent part of the immune response and also that broad inhibition of CIDRα1 proteins may be achieved through an optimal choice of multiple variants to be combined in a polyvalent vaccine strategy. The immunization with several CIDRα1.1 variants indicated that inhibition of variants not included as immunogens increased differently with addition of the different variants. Combining immunogens appeared to have little impact on elicitation of inhibitory antibodies, but without an understanding of which epitopes can confer inhibition, it is difficult to determine the number of variants required for coverage of the CIDRα1 protein family. While it is premature to speculate on the exact number of variants required, reassessment of the single CIDRα1.1 immunizations suggests that a different immunogen cocktail including just two select CIDRα1.1 domains may induce antibodies inhibiting all eight CIDRα1.1 variants, chosen to represent the sequence diversity of the subtype.
The present data show that antibodies cross-reactive between recombinant CIDRα1 domains are readily elicited and that cross-reactive inhibitory antibodies are rarer but can be elicited. The study provides a benchmark for future work aiming to elicit broadly reactive and inhibitory antibody responses.
MATERIALS AND METHODS
Recombinant protein production.
Proteins were produced in baculovirus-infected High Five cells as previously described (23, 29), albeit with different C-terminal tags for 30-kDa and 19-kDa CIDRα1 domains. In short, DNA encoding domains CIDRα2 to CIDRα6 and ∼30-kDa CIDRα1 domains according to the domain boundaries defined in Rask et al. (24) were synthesized (Geneart, Regensburg, Germany) and codon optimized for expression using a baculovirus-insect cell expression system. These proteins were expressed with C-terminal V5 and His tags and purified by nickel affinity chromatography. The ∼19-kDa CIDRα1 domains were produced by a baculovirus-insect cell expression system using codon-optimized synthetic DNA encoding CIDRα1 domains without the N-terminal beta-sheets but including the triple alpha-helix structure comprising the functional EPCR binding structure of CIDRα1, as defined in Lau et al. (23) (corresponding to amino acid positions 567 to 719 in the HB3var03 reference gene sequence). The ∼19-kDa CIDRα1 domains were produced as Strep-tag II fusion proteins and purified on a StrepTrap high-performance (HP) column (GE Healthcare Life Sciences).
Immunizations and IgG purification and depletion.
Rats were immunized subcutaneously three times, at 3-week intervals, with 10 μg of each protein per rat per immunization. Freund's incomplete adjuvant was used for all immunizations. Two weeks after the last immunization, plasma was collected for analysis. Approval for the study was granted by the Animal Experiments Inspectorate of Denmark. IgG was purified using GammaBind Plus Sepharose (BD Biosciences) according to the manufacturer's protocol. Relevant IgG was then further depleted for reactivity to a V5-His peptide to remove tag-specific IgG. In brief, a HiTrap N-hydroxysuccinimide (NHS)-activated HP affinity column (GE Healthcare Life Sciences) was coupled with 4 mg of V5-His peptide according to the manufacturer's protocol. The IgG was passed over the column seven times, with the run-through collected each time. The depleted IgG was dialyzed in phosphate-buffered saline (PBS) overnight and concentrated to 1 mg/ml using a spin column.
ELISA and Luminex.
V5-His depletion was confirmed by testing enzyme-linked immunosorbent assay (ELISA) recognition to a panel of 30 different Duffy-like binding domains (DBLs) produced in an identical way as the 30-kDa CIDR (cysteine-rich interdomain region) domains (30). Background reactivity was calculated as means +2 standard deviations (SD) of each IgG sample to all 30 domains. Recognition and inhibition were tested by ELISA as described by Lau et al. (23) and by Luminex as described by Cham et al. (30). In short, for recognition ELISA, proteins were coated at 5 μg/ml, and serum was diluted to 1:100. For the Luminex assay, serum was diluted 1:80, and secondary phycoerythrin (PE)-conjugated antibody was diluted to 1:3,000. For EPCR binding inhibition ELISAs, 3 μg/ml EPCR was coated overnight, and EPCR binding domains were preincubated with 25% purified IgG at 1 mg/ml for 1 h prior to incubation with EPCR.
Sequence and data analysis.
CIDRα1 sequence alignments were made using MUSCLE (31), and visual inspection was performed using a BioEdit sequence alignment editor. The sequence logo was made using WebLogo (32), and data were analyzed using Microsoft Excel and STATA (StataCorp).
ACKNOWLEDGMENTS
This project was supported by funding from Lundbeck Foundation, Novo Nordisk Fonden, and Danish Council for Independent Research (DFF–4004-00624B).
We declare that we have no conflicts of interest.
We thank Susanne L. Nielsen for technical assistance in protein production and Anne Corfitz, Jens E. V. Petersen, and Christian W. Wang for assisting with immunization of animals.
REFERENCES
- 1.Hviid L, Jensen AT. 2015. PfEMP1—a parasite protein family of key importance in Plasmodium falciparum malaria immunity and pathogenesis. Adv Parasitol 88:51–84. doi: 10.1016/bs.apar.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Bernabeu M, Smith JD. 2017. EPCR and malaria severity: the center of a perfect storm. Trends Parasitol 33:295–308. doi: 10.1016/j.pt.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seydel KB, Kampondeni SD, Valim C, Potchen MJ, Milner DA, Muwalo FW, Birbeck GL, Bradley WG, Fox LL, Glover SJ, Hammond CA, Heyderman RS, Chilingulo CA, Molyneux ME, Taylor TE. 2015. Brain swelling and death in children with cerebral malaria. N Engl J Med 372:1126–1137. doi: 10.1056/NEJMoa1400116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishioka H, Ghose A, Charunwatthana P, Maude R, Plewes K, Kingston H, Intharabut B, Woodrow C, Chotivanich K, Sayeed AA, Hasan MU, Day NP, Faiz A, White NJ, Hossain A, Dondorp AM. 2016. Sequestration and red cell deformability as determinants of hyperlactatemia in falciparum malaria. J Infect Dis 213:788–793. doi: 10.1093/infdis/jiv502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakobsen PH, Morris-Jones S, Theander TG, Hviid L, Hansen MB, Bendtzen K, Ridley RG, Greenwood BM. 1994. Increased plasma levels of soluble IL-2R are associated with severe Plasmodium falciparum malaria. Clin Exp Immunol 96:98–103. doi: 10.1111/j.1365-2249.1994.tb06237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsh K, Forster D, Waruiru C, Mwangi I, Winstanley M, Marsh V, Newton C, Winstanley P, Warn P, Peshu N, Pasvol G, Snow R. 1995. Indicators of life-threatening malaria in African children. N Engl J Med 332:1399–1404. doi: 10.1056/NEJM199505253322102. [DOI] [PubMed] [Google Scholar]
- 7.von Seidlein L, Olaosebikan R, Hendriksen IC, Lee SJ, Adedoyin OT, Agbenyega T, Nguah SB, Bojang K, Deen JL, Evans J, Fanello CI, Gomes E, Pedro AJ, Kahabuka C, Karema C, Kivaya E, Maitland K, Mokuolu OA, Mtove G, Mwanga-Amumpaire J, Nadjm B, Nansumba M, Ngum WP, Onyamboko MA, Reyburn H, Sakulthaew T, Silamut K, Tshefu AK, Umulisa N, Gesase S, Day NP, White NJ, Dondorp AM. 2012. Predicting the clinical outcome of severe falciparum malaria in African children: findings from a large randomized trial. Clin Infect Dis 54:1080–1090. doi: 10.1093/cid/cis034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta S, Snow RW, Donnelly CA, Marsh K, Newbold C. 1999. Immunity to non-cerebral severe malaria is acquired after one or two infections. Nat Med 5:340–343. doi: 10.1038/6560. [DOI] [PubMed] [Google Scholar]
- 9.Goncalves BP, Huang CY, Morrison R, Holte S, Kabyemela E, Prevots DR, Fried M, Duffy PE. 2014. Parasite burden and severity of malaria in Tanzanian children. N Engl J Med 370:1799–1808. doi: 10.1056/NEJMoa1303944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bull PC, Abdi AI. 2016. The role of PfEMP1 as targets of naturally acquired immunity to childhood malaria: prospects for a vaccine. Parasitology 143:171–186. doi: 10.1017/S0031182015001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mkumbaye SI, Wang CW, Lyimo E, Jespersen JS, Manjurano A, Mosha J, Kavishe RA, Mwakalinga SB, Minja DT, Lusingu JP, Theander TG, Lavstsen T. 2017. The severity of Plasmodium falciparum infection is associated with transcript levels of var genes encoding EPCR-binding PfEMP1. Infect Immun 85:e00841-16. doi: 10.1128/IAI.00841-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shabani E, Hanisch B, Opoka RO, Lavstsen T, John CC. 2017. Plasmodium falciparum EPCR-binding PfEMP1 expression increases with malaria disease severity and is elevated in retinopathy negative cerebral malaria. BMC Med 15:183. doi: 10.1186/s12916-017-0945-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuikue Ndam N, Moussiliou A, Lavstsen T, Kamaliddin C, Jensen ATR, Mama A, Tahar R, Wang CW, Jespersen JS, Alao JM, Gamain B, Theander TG, Deloron P. 2017. Parasites causing cerebral falciparum malaria bind multiple endothelial receptors and express EPCR and ICAM-1-binding PfEMP1. J Infect Dis 215:1918–1925. doi: 10.1093/infdis/jix230. [DOI] [PubMed] [Google Scholar]
- 14.Jespersen JS, Wang CW, Mkumbaye SI, Minja DT, Petersen B, Turner L, Petersen JE, Lusingu JP, Theander TG, Lavstsen T. 2016. Plasmodium falciparum var genes expressed in children with severe malaria encode CIDRα1 domains. EMBO Mol Med 8:839–850. doi: 10.15252/emmm.201606188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shabani E, Opoka RO, Bangirana P, Park GS, Vercellotti GM, Guan W, Hodges JS, Lavstsen T, John CC. 2016. The endothelial protein C receptor rs867186-GG genotype is associated with increased soluble EPCR and could mediate protection against severe malaria. Sci Rep 6:27084. doi: 10.1038/srep27084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertin GI, Lavstsen T, Guillonneau F, Doritchamou J, Wang CW, Jespersen JS, Ezimegnon S, Fievet N, Alao MJ, Lalya F, Massougbodji A, Ndam NT, Theander TG, Deloron P. 2013. Expression of the domain cassette 8 Plasmodium falciparum erythrocyte membrane protein 1 is associated with cerebral malaria in Benin. PLoS One 8:e68368. doi: 10.1371/journal.pone.0068368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavstsen T, Turner L, Saguti F, Magistrado P, Rask TS, Jespersen JS, Wang CW, Berger SS, Baraka V, Marquard AM, Seguin-Orlando A, Willerslev E, Gilbert MT, Lusingu J, Theander TG. 2012. Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proc Natl Acad Sci U S A 109:E1791–E1800. doi: 10.1073/pnas.1120455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magallon-Tejada A, Machevo S, Cistero P, Lavstsen T, Aide P, Rubio M, Jimenez A, Turner L, Valmaseda A, Gupta H, De Las Salas B, Mandomando I, Wang CW, Petersen JE, Munoz J, Gascon J, Macete E, Alonso PL, Chitnis CE, Bassat Q, Mayor A. 2016. Cytoadhesion to gC1qR through Plasmodium falciparum erythrocyte membrane protein 1 in severe malaria. PLoS Pathog 12:e1006011. doi: 10.1371/journal.ppat.1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdi AI, Kariuki SM, Muthui MK, Kivisi CA, Fegan G, Gitau E, Newton CR, Bull PC. 2015. Differential Plasmodium falciparum surface antigen expression among children with malarial retinopathy. Sci Rep 5:18034. doi: 10.1038/srep18034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernabeu M, Danziger SA, Avril M, Vaz M, Babar PH, Brazier AJ, Herricks T, Maki JN, Pereira L, Mascarenhas A, Gomes E, Chery L, Aitchison JD, Rathod PK, Smith JD. 2016. Severe adult malaria is associated with specific PfEMP1 adhesion types and high parasite biomass. Proc Natl Acad Sci U S A 113:E3270–E3279. doi: 10.1073/pnas.1524294113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessler A, Dankwa S, Bernabeu M, Harawa V, Danziger SA, Duffy F, Kampondeni SD, Potchen MJ, Dambrauskas N, Vigdorovich V, Oliver BG, Hochman SE, Mowrey WB, MacCormick IJC, Mandala WL, Rogerson SJ, Sather DN, Aitchison JD, Taylor TE, Seydel KB, Smith JD, Kim K. 2017. Linking EPCR-binding PfEMP1 to brain swelling in pediatric cerebral malaria. Cell Host Microbe 22:601–614.e5. doi: 10.1016/j.chom.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner L, Lavstsen T, Berger SS, Wang CW, Petersen JE, Avril M, Brazier AJ, Freeth J, Jespersen JS, Nielsen MA, Magistrado P, Lusingu J, Smith JD, Higgins MK, Theander TG. 2013. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature 498:502–505. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau CK, Turner L, Jespersen JS, Lowe ED, Petersen B, Wang CW, Petersen JE, Lusingu J, Theander TG, Lavstsen T, Higgins MK. 2015. Structural conservation despite huge sequence diversity allows EPCR binding by the PfEMP1 family implicated in severe childhood malaria. Cell Host Microbe 17:118–129. doi: 10.1016/j.chom.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rask TS, Hansen DA, Theander TG, Gorm PA, Lavstsen T. 2010. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes—divide and conquer. PLoS Comput Biol 6:e1000933. doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sander AF, Lavstsen T, Rask TS, Lisby M, Salanti A, Fordyce SL, Jespersen JS, Carter R, Deitsch KW, Theander TG, Pedersen AG, Arnot DE. 2014. DNA secondary structures are associated with recombination in major Plasmodium falciparum variable surface antigen gene families. Nucleic Acids Res 42:2270–2281. doi: 10.1093/nar/gkt1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner L, Lavstsen T, Mmbando BP, Wang CW, Magistrado PA, Vestergaard LS, Ishengoma DS, Minja DT, Lusingu JP, Theander TG. 2015. IgG antibodies to endothelial protein C receptor-binding cysteine-rich interdomain region domains of Plasmodium falciparum erythrocyte membrane protein 1 are acquired early in life in individuals exposed to malaria. Infect Immun 83:3096–3103. doi: 10.1128/IAI.00271-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barfod L, Dobrilovic T, Magistrado P, Khunrae P, Viwami F, Bruun J, Dahlback M, Bernasconi NL, Fried M, John D, Duffy PE, Salanti A, Lanzavecchia A, Lim CT, Ndam NT, Higgins MK, Hviid L. 2010. Chondroitin sulfate A-adhering Plasmodium falciparum-infected erythrocytes express functionally important antibody epitopes shared by multiple variants. J Immunol 185:7553–7561. doi: 10.4049/jimmunol.1002390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ntumngia FB, Pires CV, Barnes SJ, George MT, Thomson-Luque R, Kano FS, Alves JRS, Urusova D, Pereira DB, Tolia NH, King CL, Carvalho LH, Adams JH. 2017. An engineered vaccine of the Plasmodium vivax Duffy binding protein enhances induction of broadly neutralizing antibodies. Sci Rep 7:13779. doi: 10.1038/s41598-017-13891-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsieh FL, Turner L, Bolla JR, Robinson CV, Lavstsen T, Higgins MK. 2016. The structural basis for CD36 binding by the malaria parasite. Nat Commun 7:12837. doi: 10.1038/ncomms12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cham GK, Turner L, Lusingu J, Vestergaard L, Mmbando BP, Kurtis JD, Jensen AT, Salanti A, Lavstsen T, Theander TG. 2009. Sequential, ordered acquisition of antibodies to Plasmodium falciparum erythrocyte membrane protein 1 domains. J Immunol 183:3356–3363. doi: 10.4049/jimmunol.0901331. [DOI] [PubMed] [Google Scholar]
- 31.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]