Brucellaceae are a group of pathogenic intracellular bacteria with the ability to modulate the host response, both at the individual cell level and systemically. One of the hallmarks of the virulence process is the capacity of the bacteria to downregulate the adaptive and acquired host immune response through a plethora of virulence factors that directly impact several key signaling cascades.
KEYWORDS: Brucella, effector protein, palmitoylation
ABSTRACT
Brucellaceae are a group of pathogenic intracellular bacteria with the ability to modulate the host response, both at the individual cell level and systemically. One of the hallmarks of the virulence process is the capacity of the bacteria to downregulate the adaptive and acquired host immune response through a plethora of virulence factors that directly impact several key signaling cascades. PrpA is one of those virulence factors that alters, via its polyclonal B-cell activity, the humoral and cellular immune responses of the host, ultimately favoring the establishment of a chronic infection. Even though PrpA affects B cells, it directly targets macrophages, triggering a response that ultimately affects B lymphocytes. In the present article we report that PrpA is S-palmitoylated in two N-terminal cysteine residues by the host cell and that this modification is necessary for its biological activity. Our results demonstrate that S-palmitoylation promotes PrpA migration to the host cell plasma membrane and stabilizes the protein during infection. These findings add a new mechanism exploited by this highly evolved pathogen to modulate the host immune response.
INTRODUCTION
Brucellaceae are widespread zoonotic intracellular pathogens that cause brucellosis, a disease that inflicts important economic losses in animal production since it induces premature abortion of pregnant heifers. Because of its zoonotic potential (transmitted by either contact with infected animals or consumption of nonpasteurized dairy products), brucellosis is still a human health concern, particularly in areas of endemicity (1, 2).
Brucella abortus is a chronic pathogen that has evolved a wide variety of immune evasion strategies in order to persist in the context of a robust immune response. These strategies include downregulating the activation of certain pathogen-associated molecular patterns (PAMPs), synthesis of structural components with low proinflammatory activities, and alteration of the normal course of the adaptive immune response (3, 4). One example of the last type of modulation is the one triggered by PrpA (for proline racemase protein A), a protein of Brucella that our group has identified and characterized as a polyclonal B-cell mitogen involved in the establishment of the chronic phase of the infectious process in mice (5). PrpA is a strong immune modulator as it induces B-cell proliferation in vitro and in vivo, T-cell anergy, and alteration of the cytokine pattern and humoral immune response during infection (6). Interestingly, even though PrpA induces polyclonal B-cell proliferation, its cellular target is actually macrophages. Our working model is that intramacrophagic Brucella organisms are able to secrete and translocate PrpA from the Brucella-containing vacuole (BCV), which traffics to the plasma membrane where it engages its receptor to trigger the secretion of one or more soluble factors that ultimately induce B-cell proliferation (7). This model implies that PrpA must have specific mechanisms to reach the cellular membrane in order to engage its receptor(s) (7).
In the present report we have further advanced in understanding how PrpA reaches the host cell membrane. We show that this protein is palmitoylated in two cysteine residues of the N-terminal region and that this posttranslational modification is necessary for its localization to the plasma membrane in transfection assays. We show by immunofluorescence that in cell culture PrpA is secreted from Brucella and translocated through the BCV and that mutagenesis of these amino acids results in a protein that is rapidly degraded by the host cell. Moreover, the function of PrpA during infection is also altered when these two cysteine residues are eliminated, indicating that the biological activities of PrpA are palmitoylation dependent. Our results indicate that the host-mediated S-palmitoylation of this virulence factor can alter its stability, its localization, and ultimately its biological function.
RESULTS
PrpA is exposed in the plasma membrane of the eukaryotic cell, and this localization is dependent on its palmitoylation status.
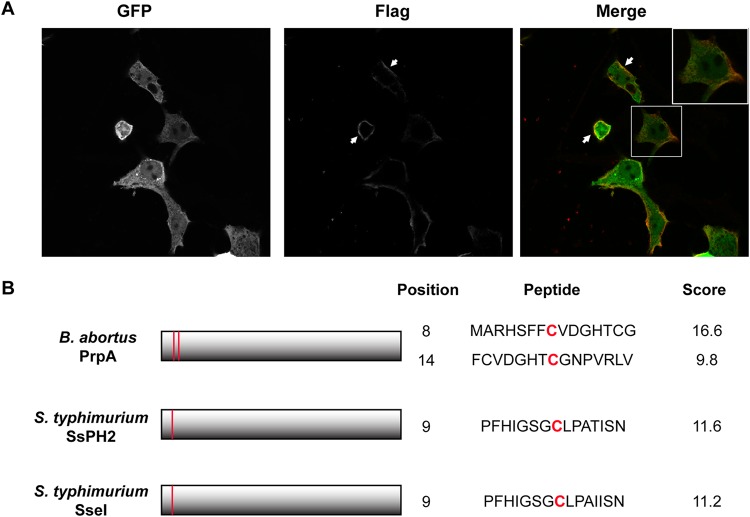
Our current working hypothesis is that during infection, PrpA translocates across the Brucella-containing-vacuole (BCV), traffics to the plasma membrane of the host cell, and finally is exposed on the surface, where it engages its receptor(s) (7). To test this possibility, we transfected HEK293 cells with a eukaryotic expression vector coding for PrpA fused to green fluorescent protein (GFP) and a 3×Flag tag (PrpA-GFP) and analyzed the localization of the fusion protein by confocal microscopy. To determine if PrpA-GFP reaches the surface of the cell, we performed immunofluorescence staining in nonpermeabilized cells with a monoclonal anti-Flag antibody to determine its localization. As can be observed in Fig. 1A, PrpA was detected on the surface of nonpermeabilized HEK293 cells, indicating that PrpA hijacks specific cellular components and traffics to the outer face of the cell membrane. Since PrpA traffics and is retained in the host cell membrane, we searched the sequence of the protein for potential motifs that might promote this. S-palmitoylation is a eukaryotic posttranslational modification consisting of a reversible covalent attachment of a palmitoyl chain to cysteine residues by a thioester bond. Besides affecting localization and function, S-palmitoylation has been shown to promote migration and retention of proteins in membranes. We found that PrpA has two cysteine residues (residues 8 and 14) in its N-terminal region, with a predictive high score for this modification using the high-performance protein palmitoylation site predictor program CSS-Palm 4.0 (Fig. 1B).
FIG 1.
PrpA is translocated to the cell membrane. (A) HEK293 cells were transfected with pEGFP-PrpA3×Flag and examined by indirect immunofluorescence confocal microscopy using an anti-Flag monoclonal antibody under nonpermeabilization conditions. Green, GFP; red, Flag. (B) Schematic representation of B. abortus PrpA and S. enterica serovar Typhimurium SseI and SspH2, highlighting the predicted palmitoylation motifs and their scores.
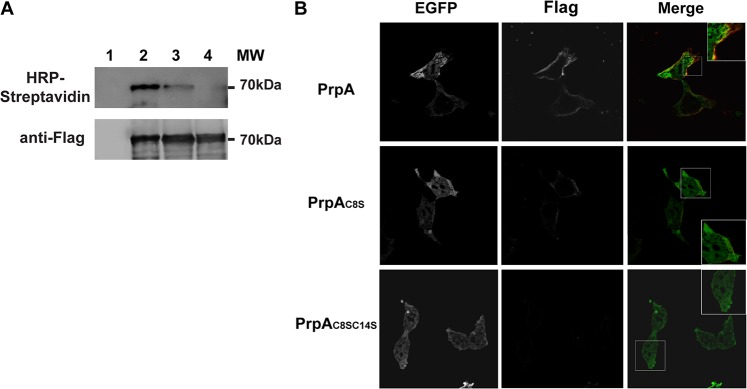
To directly test whether PrpA is palmitoylated, we utilized bio-orthogonal labeling to detect the incorporation of an alkylated analogue of palmitic acid (17-octadecynoic acid [17-ODYA]). HEK293 cells transfected with the construct that expresses the PrpA-GFP-3×Flag fusion were metabolically labeled with 17-ODYA for 3 h, lysed, immunoprecipitated with a monoclonal anti-Flag antibody, and subjected to a click-it reaction with biotin-azide. In this step, the chemoselective ligation, or “click-it” reaction, between the biotin-azide and the alkyne group of the 17-octadecynoic acid occurs, and thus metabolically 17-ODYA-labeled proteins become biotinylated. S-palmitoylation was finally detected by SDS-PAGE and Western blotting with a streptavidin-horseradish peroxidase (HRP)-conjugated antibody (see Materials and Methods for a complete description of the protocol). Figure 2A shows that PrpA was palmitoylated and that this modification was abolished when both cysteine residues 8 and 14 were mutated to serine. As shown in the anti-Flag antibody panel, the stabilities of all constructs were similar.
FIG 2.
S-palmitoylation of cysteine residues 8 and 14 is required for PrpA membrane localization. (A) HEK293 cells were transfected with pEGFP, pEGFP-PrpA3×Flag, pEGFP-PrpAC8S3×Flag, or pEGFP-PrpAC8SC14S-3×Flag (lanes 1, 2, 3, and 4, respectively) and metabolically labeled with 17-ODYA. Labeled cells were lysed, and PrpA fusion proteins were immunoprecipitated with a monoclonal anti-Flag antibody. Click-it chemistry was performed on the immunoprecipitates using biotin-azide, followed by SDS-PAGE and immunoblotting with streptavidin-HRP to detect palmitoylation and anti-Flag antibody as a loading control. (B) Transfected HEK293 cells (green) were examined by immunofluorescence confocal microscopy using an antibody directed to the Flag epitope (red) under nonpermeabilization conditions.
To determine if PrpA S-palmitoylation is necessary for membrane association, we analyzed by immunofluorescence and confocal microscopy the localization of the PrpA-GFP-3×Flag double mutant (PrpAC8SC14S) in transfected HEK293 cells. Figure 2B shows that the double-point-mutation construct lost its membrane localization, indicating that S-palmitoylation is necessary for the proper trafficking and association of PrpA with the plasma membrane.
Palmitoylation is necessary for PrpA immunomodulatory functions in vivo.
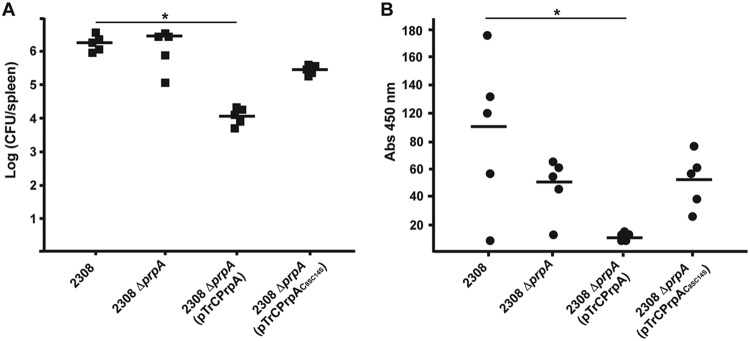
We have previously shown that PrpA is a strong immunomodulator that affects virulence and anti-Brucella immunoglobulin titers during Brucella infection in mice (6). To determine whether these PrpA immunomodulatory activities are S-palmitoylation dependent, we analyzed the capacity of PrpAC8SC14S to complement the B. abortus prpA deletion mutant in vivo. We constructed a PrpA-3×Flag or PrpAC8SC14S-3×Flag fusion protein expressed from the strong promoter of plasmid pTrc, which has been shown to have high levels of expression in Brucella, and used them to complement the B. abortus prpA mutant strain. Surprisingly, overexpression of PrpA in the mutant resulted in a strain with reduced fitness in mice (Fig. 3A), indicating that high levels of the immunomodulatory activity are detrimental for the infectious process. On the other hand, overexpression of PrpAC8SC14S had no statistically significant effect compared to the case for the wild type or the prpA mutant, indicating a direct link between biological activity and palmitoylation. Consistent with this observation, the decrease in the anti-Brucella IgG antibody titers observed in the PrpA mutant was actually more pronounced in the strain overexpressing PrpA, while overexpression of PrpAC8SC14S again had no effect (Fig. 3B). These results indicate that palmitoylation sites are required for PrpA immunomodulatory activities in vivo.
FIG 3.
Palmitoylation sites are required for PrpA biological activity in mice. BALB/c mice were experimentally infected with the Brucella abortus strains described in the text and sacrificed at 10 days postinfection. (A) Bacterial loads in the spleens were determined by dilution and CFU quantification. *, P < 0.01. (B) Anti-Brucella specific antibody titers in serum samples from the infected mice were determined by ELISA. *, P < 0.05.
S-palmitoylation of PrpA stabilizes the protein during infection.
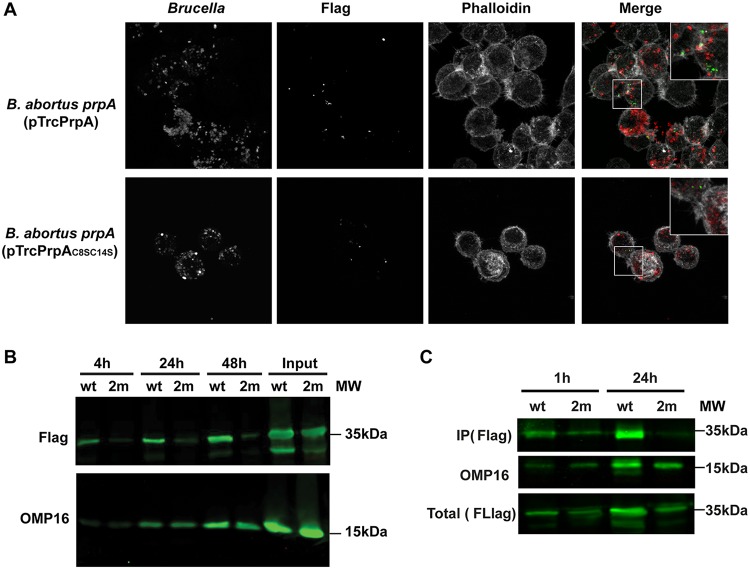
The demonstration that S-palmitoylation sites are necessary for PrpA biological activity led us to evaluate and compare the secretion and translocation of PrpA and PrpAC8SC14S during the infection of host cells. In order to study this, J774 A.1 cells were infected with B. abortus prpA(pTrC-PrpA-3×Flag) or B. abortus prpA(pTrC-PrpAC8SC14S-3×Flag), and translocation was detected by immunofluorescence confocal microscopy. Since the protocol for immunostaining causes permeabilization of infected cells but not bacteria (8), detection of the Flag signal implies that PrpA is secreted from Brucella. Figure 4A shows that, at 4 and 24 h postinfection, PrpA is secreted from the bacteria and localizes in the vicinity of Brucella in discrete foci. Surprisingly, we were unable to detect PrpAC8SC14S either by immunofluorescence or by Western blotting (Fig. 4A and B). The level of infection of both strains was the same as evidenced by Western blotting using a monoclonal antibody against outer membrane protein 16 (OMP16) of Brucella (Fig. 4B). Additionally, the stabilities of both constructs during axenic growth in vitro (Fig. 4B) and in transfection assays (Fig. 2) were equivalent, indicating that the instability of PrpAC8SC14S occurs only during infection. In order to analyze whether PrpAC8SC14S is degraded before or after the translocation step, J774 A.1 cells were infected with the B. abortus prpA(pTrC-PrpA-3×Flag) or B. abortus prpA(pTrC-PrpAC8SC14S-3×Flag) strain, and at 1 h or 24 h postinfection, the cells were lysed in Brij buffer with 1% SDS, centrifuged, and filtered in order to remove debris as well as intracellular bacteria. This supernatant, which contained the cytoplasmic soluble fraction of the infected cells, was immunoprecipitated with an anti-Flag-M2 antibody to detect PrpA translocation. As can be observed in Fig. 4C, PrpAC8SC14S was detected at levels similar to those of PrpA in the cytoplasmic fraction of the infected cells at 1 h but not at 24 h postinfection, indicating that PrpA is translocated early after internalization of the bacteria and degraded during the infectious process. These results demonstrate that PrpA S-palmitoylation stabilizes the protein in the host cell and strongly suggest that the lack of activity observed in the in vivo experiments is probably due to degradation of the protein in infected cells.
FIG 4.
S-palmitoylation is required for PrpA stability during infection. J774 A.1 cells were infected with the Brucella abortus prpA mutant strain complemented with PrpA-3×Flag (wild type [wt]) or PrpAC8SC14S-3×Flag (2m). (A) At 24 h postinfection, the cells were fixed and examined by immunofluorescence confocal microscopy using anti-Brucella antibodies (red), an anti-Flag monoclonal antibody (green), and phalloidin to stain polymerized actin (white). (B) At 4, 24, and 48 h postinfection, the infected cells were subjected to SDS-PAGE and immunoblotting with an anti-Flag monoclonal antibody or an anti-OMP16 antibody as a loading control. Input is the in vitro axenic culture used to infect cells. (C) At 1 and 24 h postinfection, cells were lysed, centrifuged, and filtered to remove intracellular bacteria. PrpA-3×Flag (wt) or PrpAC8SC14S-3×Flag (2m) was immunoprecipitated with a anti-Flag monoclonal antibody, subjected to SDS-PAGE, and immunoblotted with an anti-Flag monoclonal antibody.
DISCUSSION
Many pathogenic bacteria have the ability to inject or translocate into the host cells effector proteins that alter different cellular activities to promote pathogenicity. These virulence factors need to be stabilized and localized in specific compartments of the cell to find and modulate their targets (9). Brucella is an intracellular pathogen with the capacity to invade and replicate in many cell types of the infected host and to establish a persistent infection (3, 4). To achieve this, the bacterium has the capacity to translocate to the host cells a plethora of proteins that alter either the normal trafficking of the intracellular bacteria or the immune response triggered by the infection (3, 10). PrpA is one of these effector proteins, and we have previously shown that this virulence factor is involved in the modulation of the immune response, acting on macrophages and triggering a polyclonal B-cell proliferation response that allows the bacterium to establish a persistent infection (5–7). How the protein is targeted to the plasma membrane of the host cell after its translocation from the Brucella-containing vacuole has not been elucidated until now. In the present work, we demonstrate that PrpA is translocated to the cell membrane in transfection assays and that S-palmitoylation of two amino-terminal cysteine residues is necessary for this localization. We further show that the proper activity of PrpA is dependent on its S-palmitoylation status, indicating that this host cell modification is necessary for its function. Our results indicate that during infection, stabilization of PrpA is dependent on the S-palmitoylation of these cysteine residues.
S-palmitoylation is a eukaryotic posttranslational modification that consists, in general, of the covalent attachment of a saturated 16-carbon palmitic acid to a cysteine residue of the target protein through a thioester bond. This modification has been shown to play an important role in regulating protein activity, stability, and localization (12). Our results demonstrate that Brucella abortus hijacks this eukaryotic posttranslational process, exploiting host biology in its own favor. S-palmitoylation of four bacterial effectors during infection has been already described. Salmonella enterica type III secreted effectors SspH2 and Sse1 localize to the plasma membrane of the infected cell, and that localization is dependent on the S-palmitoylation of a cysteine residue in their N-terminal regions (13). GobX is a type IV secreted effector of Legionella pneumophila that also requires host cell palmitoylation of a cysteine residue for its specific localization (14). L. pneumophila has a second effector protein that is palmitoylated, LpdA, a virulence factor with phospholipase D activity (15).
Effector stability is central during the intracellular infectious cycle and has to be finely tuned by the pathogen in order to modulate the host response. Ubiquitination has been shown to be a common modification used by these virulence factors to stabilize or alter the half-lives of the proteins once they are translocated to the host cell (9, 16–18). To our knowledge, this is the first report of a bacterial effector protein whose stability in the host cell is dependent on S-palmitoylation. Is this modification required for PrpA stability or for membrane localization, which ultimately stabilizes the protein? The latter seems not to be the case because PrpAC8SC14S does not localize to the plasma membrane and is still stable during cell transfection experiments. Our working model is that S-palmitoylation stabilizes translocated PrpA and promotes its migration to the host cell plasmatic membrane where it engages its receptor, triggering a macrophagic response that ultimately promotes B-cell proliferation.
MATERIALS AND METHODS
Bacterial strains, expression constructs, and cell lines.
Brucella abortus 2308 was used as a wild-type strain. B. abortus strains were grown in tryptic soy agar (Difco/BD Biosciences) or in tryptic soy broth (TSB) at 37°C on a rotary shaker for 16 to 24 h. Manipulation of B. abortus was performed at the biosafety level 3 (BSL3) laboratory facility at the Universidad Nacional de San Martín. If necessary, medium was supplemented with antibiotics at the following final concentrations: ampicillin, 100 μg/ml; and nalidixic acid, 5 μg/ml. For eukaryotic expression, full-length PrpA was amplified from the Brucella melitensis bv. abortus 2308 genome and cloned in vector pEGFP-N1 (Addgene) in frame with a C-terminal 3×Flag tag. PrpAC8SC14S was created using 5′ primers designed with a point mutation changing the cysteine codons (TGC) at positions 8 and 14 of PrpA to serine (AGC). For bacterial expression, full-length PrpA-3×Flag and PrpAC8SC14S-3×Flag were subcloned from pEGFP-N1 to the high-expression vector pTrC (19), generating the pTrcPrpA-3×Flag or pTrcPrpAC8SC14S-3×Flag plasmid. These plasmids were introduced to complement the Brucella abortus prpA deletion mutant, generating prpA(pTrcPrpA) and prpA(pTrcPrpaC8SC14S) Brucella abortus strains. Protein size and stability in vitro, in transfected cells, and in infected cells were determined by Western blotting using mouse anti-Flag M2 monoclonal antibody (1:5,000; Sigma) and IRDye secondary anti-mouse antibody (1:20,000; Li-Cor Inc.). All antibodies were diluted in a solution of Tris-buffered saline (TBS), 1% nonfat milk, and 0.1% Tween. Detection was performed using the Odyssey imaging system (Li-Cor, Inc.). HeLa, HEK293, and J774 A.1 cells were maintained in Dulbecco modified Eagle medium (DMEM) (Invitrogen) containing 5% fetal bovine serum (FBS) at 37°C in 5% CO2.
Cell transfection and immunofluorescence assays.
HEK293 cells were seeded on 12-mm coverslips in 24-well plates at 5 × 104 cells per well and transfected with pEGFP-PrpA3×Flag or pEGFP-PrpAC8SC14S3×Flag using Lipofectamine 3000 (Invitrogen), according to the manufacturer's instructions. At 24 h posttransfection, cells were washed three times with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (pH 7.4) for 15 min at 37°C. For localization studies, all incubations were performed under nonpermeabilizing conditions. Fixed cells were washed twice and coverslips incubated for 30 min in blocking buffer containing primary antibodies. After two washes in PBS, the coverslips were incubated for 30 min in blocking buffer containing secondary antibodies. Finally, the coverslips were washed three times in PBS and once in Milli-Q water and mounted on glass slides using Fluorsave (Calbiochem). The primary antibody used was M2 mouse anti-Flag (Sigma) at 1:4,000. The secondary antibody used was Alexa Fluor 568 goat anti-mouse IgG (Molecular Probes, Invitrogen) at 1:4,000. Confocal images were acquired using an IX-81 microscope attached to a FV-1000 confocal module, with a Plan APO 60×, 1.42-numerical-aperture (NA) oil immersion objective (Olympus, Japan). The acquisition software used was FV 10-ASW 3.1. Images were treated using ImageJ 1.45s software (NIH, USA), and images of 1,024 by 1,024 pixels were then assembled using ImageJ software.
In vitro palmitoylation assay.
In vitro palmitoylation experiments were carried out as described in reference 13. HEK293 cells were seeded in a P100 culture plate until 50% to 60% confluence and transfected with pEGFP-PrpA3×Flag or pEGFP-PrpAC8SC14S-3×Flag as described above. At 24 h posttransfection, cells were washed with PBS, starved for 60 min in DMEM, and metabolically labeled with dimethyl sulfoxide (DMSO) or 17-ODYA (100 mg/ml) in DMEM with 1% bovine serum albumin (BSA) for 3 h. Cells were then washed 3 times with PBS and lysed in Brij lysis buffer (1% Brij 97, 150 mM NaCl, 50 mM triethanolamine, pH 7.4) with EDTA-free protease inhibitor cocktail on ice. Cell lysates were collected following centrifuging at 14,000 rpm for 20 min at 4°C to remove debris. Immunoprecipitations were performed using anti-Flag-M2 affinity resin (Sigma). The immunoprecipitates were washed three times with 1 ml of ice-cold modified radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 0.1% SDS, 50 mM triethanolamine [pH 7.4], 150 mM NaCl), resuspended in 50 mM Tris-HCl (pH 8.0) and 1% SDS, and subjected to click-it chemistry using biotin-azide (Invitrogen) and the Click-iT protein reaction buffer kit (Invitrogen) according to the manufacturer's instructions. Samples were then boiled in Laemmli buffer, subjected to SDS-PAGE, and detected by Western blotting using HRP-streptavidin (eBioscience) at a 1:2,500 dilution in PBS with 5% BSA.
Infection of mice and cells.
Mouse infections were carried out as described previously (5, 11). Briefly, female 60- to 90-day-old BALB/c mice were injected intraperitoneally with 0.2 ml of PBS containing 105 CFU of the wild-type B. abortus, B. abortus prpA, B. abortus prpA(pTrCPrpA), or B. abortus prpA(pTrcPrpAC8SC14S) strain. At 10 days postinfection, animals were sacrificed, and the spleens were removed, homogenized in PBS, and processed for direct CFU determination (plating). All mice were bred in accordance with institutional animal guidelines under specific-pathogen-free conditions in the local animal facility (BSL3; Institute for Research in Biotechnology) of the University of San Martín. Mouse studies were approved by the local regulatory agencies (CICUAE-UNSAM). For PrpA secretion analysis, J774 A.1 cells were infected and staining performed as described in reference 8. Cells (5 × 104/well) were seeded on 24-well plates in medium without antibiotics at 24 h before infection. B. abortus infections were carried out at the a multiplicity of infection (MOI) of 500:1. Bacteria were centrifuged onto cells at 400 × g for 10 min. After 40 min, the wells were gently washed three times with phosphate-buffered saline (PBS) and incubated for 60 min with fresh medium containing 50 μg/ml gentamicin and 50 μg/ml streptomycin to kill noninternalized bacteria. Thereafter, antibiotic concentrations were decreased to 20 μg/ml gentamicin. At different times postinfection, cells were washed three times with PBS and either fixed in 4% paraformaldehyde (PFA) for immunofluorescence staining or boiled in 50 μl of Laemmli buffer and subjected to SDS-PAGE and Western blotting for detection of PrpA and PrpAC8SC14S using mouse monoclonal M2 anti-Flag antibody (Sigma). For protein secretion during infection, immunofluorescence was performed using rabbit anti-Brucella polyclonal antibody (1:1,000) and anti-Flag M2 monoclonal antibody (1:4,000). The secondary antibodies used were goat anti-mouse–Alexa Fluor 488 and goat anti-rabbit–Alexa Fluor 568 (Molecular Probes, Invitrogen) at a 1:4,000 dilution.
For immunoprecipitation assays, infected J774 A.1 cells were washed 3 times with PBS, lysed in 1 ml of Brij buffer with 1% SDS, centrifuged at 10,000 rpm, and filtered to remove debris and intracellular bacteria. Supernatants were then diluted 1:10 with PBS and immunoprecipitated using 2 μl of anti-Flag M2 antibody (Sigma) and 30 μl of 4-Fastflow-Protein G-Sepharose (GE Healthcare). Elution of immunoprecipitates was carried out in 40 μl of Laemmli buffer.
Immunoglobulin quantification.
The titers of specific immunoglobulins against Brucella were determined by indirect enzyme-linked immunosorbent assay (ELISA) (6). Briefly, ELISA MaxiSorp plates (Nunc, USA) were sensitized with 0.4 μg/well of Brucella abortus total protein extracts overnight and blocked for 2 h with 1% BSA in PBS. Serum samples from mice at 10 days postinfection were serially diluted, and total immunoglobulins were detected with HRP-conjugated secondary antibodies in a colorimetric reaction and read at 450 nm in a Benchmark microplate reader (Bio-Rad). The closest absorbance value (at 450 nm) to 0.5 was multiplied by the dilution factor to obtain the titer.
Statistical analysis.
The differences between the groups were calculated by using the Student t test for normally distributed variables and the nonparametric Mann-Whitney test for nonnormally distributed variables. A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank members of the J.E.U. laboratory for useful discussions and J. J. Letesson for the anti-OMP16 monoclonal antibody.
This work was supported by grants PICT-1028-2014 and PICT-1094-2014 to J.E.U. and J.M.S., respectively.
All authors are members of the National Research Council of Argentina (CONICET).
REFERENCES
- 1.Corbel MJ. 1997. Brucellosis: an overview. Emerg Infect Dis 3:213–221. doi: 10.3201/eid0302.970219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pappas G, Akritidis N, Bosilkovski M, Tsianos E. 2005. Brucellosis. N Engl J Med 352:2325–2336. doi: 10.1056/NEJMra050570. [DOI] [PubMed] [Google Scholar]
- 3.Byndloss MX, Tsolis RM. 2016. Brucella spp. Virulence factors and immunity. Annu Rev Anim Biosci 4:111–127. doi: 10.1146/annurev-animal-021815-111326. [DOI] [PubMed] [Google Scholar]
- 4.Byndloss MX, Tsolis RM. 2016. Chronic bacterial pathogens: mechanisms of persistence. Microbiol Spectr 4. doi: 10.1128/microbiolspec.VMBF-0020-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spera JM, Ugalde JE, Mucci J, Comerci DJ, Ugalde RA. 2006. A B lymphocyte mitogen is a Brucella abortus virulence factor required for persistent infection. Proc Natl Acad Sci U S A 103:16514–16519. doi: 10.1073/pnas.0603362103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spera JM, Comerci DJ, Ugalde JE. 2014. Brucella alters the immune response in a prpA-dependent manner. Microb Pathog 67–68:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spera JM, Herrmann CK, Roset MS, Comerci DJ, Ugalde JE. 2013. A Brucella virulence factor targets macrophages to trigger B-cell proliferation. J Biol Chem 288:20208–20216. doi: 10.1074/jbc.M113.453282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dohmer PH, Valguarnera E, Czibener C, Ugalde JE. 2014. Identification of a type IV secretion substrate of Brucella abortus that participates in the early stages of intracellular survival. Cell Microbiol 16:396–410. doi: 10.1111/cmi.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hicks SW, Galan JE. 2013. Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat Rev Microbiol 11:316–326. doi: 10.1038/nrmicro3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ke Y, Wang Y, Li W, Chen Z. 2015. Type IV secretion system of Brucella spp. and its effectors. Front Cell Infect Microbiol 5:72. doi: 10.3389/fcimb.2015.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ugalde JE, Comerci DJ, Leguizamon MS, Ugalde RA. 2003. Evaluation of Brucella abortus phosphoglucomutase (pgm) mutant as a new live rough-phenotype vaccine. Infect Immun 71:6264–6269. doi: 10.1128/IAI.71.11.6264-6269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linder ME, Deschenes RJ. 2007. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol 8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 13.Hicks SW, Charron G, Hang HC, Galan JE. 2011. Subcellular targeting of Salmonella virulence proteins by host-mediated S-palmitoylation. Cell Host Microbe 10:9–20. doi: 10.1016/j.chom.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YH, Doms AG, Cheng E, Kim B, Evans TR, Machner MP. 2015. Host cell-catalyzed S-palmitoylation mediates Golgi targeting of the Legionella ubiquitin ligase GobX. J Biol Chem 290:25766–25781. doi: 10.1074/jbc.M115.637397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroeder GN, Aurass P, Oates CV, Tate EW, Hartland EL, Flieger A, Frankel G. 2015. Legionella pneumophila effector LpdA is a palmitoylated phospholipase D virulence factor. Infect Immun 83:3989–4002. doi: 10.1128/IAI.00785-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaus K, Hentschke M, Czymmeck N, Novikova L, Trulzsch K, Valentin-Weigand P, Aepfelbacher M, Ruckdeschel K. 2011. Destabilization of YopE by the ubiquitin-proteasome pathway fine-tunes Yop delivery into host cells and facilitates systemic spread of Yersinia enterocolitica in host lymphoid tissue. Infect Immun 79:1166–1175. doi: 10.1128/IAI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubori T, Galan JE. 2003. Temporal regulation of salmonella virulence effector function by proteasome-dependent protein degradation. Cell 115:333–342. doi: 10.1016/S0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- 18.Kubori T, Shinzawa N, Kanuka H, Nagai H. 2010. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog 6:e1001216. doi: 10.1371/journal.ppat.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guidolin LS, Morrone Seijo SM, Guaimas FF, Comerci DJ, Ciocchini AE. 2015. Interaction network and localization of Brucella abortus membrane proteins involved in the synthesis, transport, and succinylation of cyclic beta-1,2-glucans. J Bacteriol 197:1640–1648. doi: 10.1128/JB.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]