Figure 2.
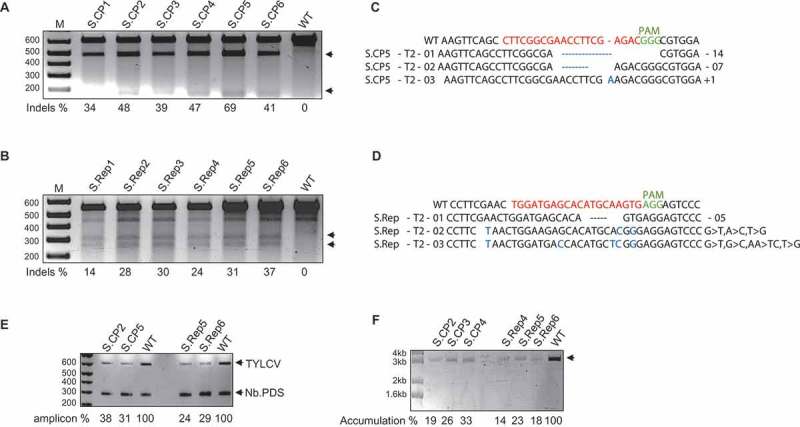
The stably-engineered CRISPR/Cas9 system conferred interference against TYLCV in tomato.TYLCV targeting by CRISPR/Cas9 in tomato plants. The T7EI assay detected InDels at the CP sequence (A) and at the Rep sequence (B) in the TYLCV PCR amplicons from plants expressing Cas9 and sgRNA, but not in the wild type used as the control. Arrows indicate the two T7EI digested fragments. (C) and (D) Alignment of reads from the PCR amplicons encompassing the CP and Rep regions of TYLCV. Top line, wild-type (WT) TYLCV sequences. InDels, are indicated by the numbers [–, deletion. +, insertion. >, substitution of x to y nucleotides]. E) Semi-quantitative PCR for the accumulation of TYLCV. A 500 bp fragment of TYLCV was amplified (22 cycles) by PCR with TYLCV-specific primers and resolved on a 1% EtBr agarose gel. N. benthamiana NBPDS-specific primers were used as an internal control. Amplicons in comparison to the wild type are represented as a percentage below the gel. F) RCA assay for the accumulation of the TYLCV genome in T2 lines expressing CRISPR-Cas9. Compared to the wild type, plants expressing CRISPR-Cas9 accumulated a reduced level of the TYLCV genome. In A, B, E, and F, arrowheads represent the expected DNA fragments.
