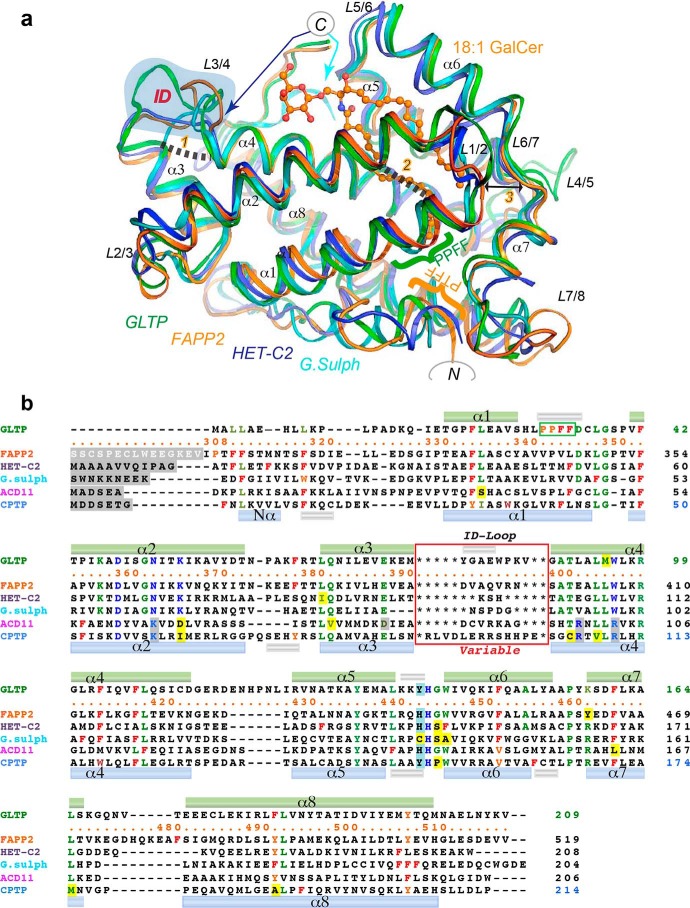
Figure 2.
Comparison of FAPP2–GLTPH domain with other family members. a, structure superimposition of the GSL-specific proteins: human FAPP2–GLTPH domain (orange), human GLTP (green), fungal heterokaryon incompatibility C2 protein, HET-C2, of P. anserina (blue), and GLTP-like protein from the thermoacidophilic unicellular red alga, G. sulfuraria (cyan) (PDB codes 5KDI, 3S0K, 4KV0, and 2I3F, respectively). 18:1-GalCer is shown as bound to FAPP2–GLTPH. The blue-shaded area highlights the ID loops. The GLTP PPFF and FAPP2 PTFF sequences are indicated by green and orange braces, respectively. Newly identified conserved linkages 1 and 2 are indicated by dashed lines; the double-headed arrow 3 points to the strictly conserved Gly and Pro on opposite sides of the gate controlling the access to the hydrophobic pocket in the GLTP fold. Colored arrows point to the C-ends of HET-C2 (blue) and G. sulfuraria protein (cyan). Color codes for GSL atoms are defined in Fig. 1. b, poststructural sequence alignment of six proteins belonging to the GLTP family (see Fig. S1a). Color codes are blue for the recognition center residues, green for the conserved/semiconserved residues, and red and orange for Phe and similarly positioned Tyr/Trp, respectively. Recognition center residues of the C1P-specific family also are blue but shaded in gray when different from the GSL-specific family. Exceptions among conserved residues are shaded by yellow. The upper and lower cylinders (small silver for 310-helices; other for α-helices) indicate the locations of secondary structure elements found in GLTP and CPTP, respectively. Within the red rectangle are the ID-loop sequences that vary among each member.