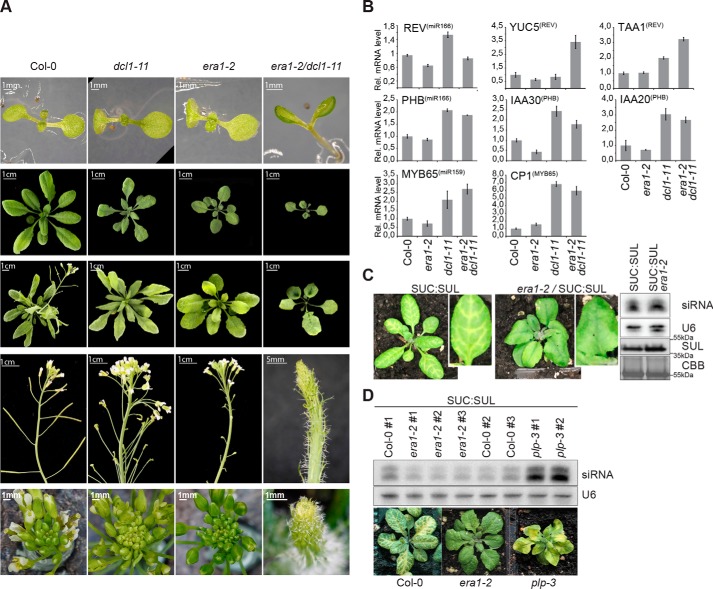
Figure 1.
Farnesyl transferase mutants show defects related to small RNA pathways. A, cotyledon, leaf, and inflorescence phenotypes of Col-0 WT, era1–2, dcl1–11, and era1–2/dcl1–11 mutants. B, relative mRNA expression levels of three miRNA targets encoding transcription factors (PHB, REV, and MYB65), as well as direct targets of these transcription factors. The figure shows results from a single biological replicate in which RNA from each genotype was prepared from pools of 12 adult leaves. Error bars indicate standard error in technical triplicates. Similar results were obtained when the entire experiment was repeated at another point in time. C, left, WT and era1–2 plants expressing the SUC:SUL (SS) hairpin. Right, ChlI (SUL) siRNA and protein levels of the individual plants shown on the left; total RNA fractions were analyzed by small RNA Northern blotting with a ChlI (SUL) probe. Total protein fractions were analyzed by Western blotting developed with ChlI antibodies. D, bottom, WT, era1–2 and plp-3 plants expressing the SUC:SUL (SS) hairpin. Top, ChlI (SUL) siRNA levels in leaves from pools of 5 plants from the F3 generation; total RNA fractions were analyzed by small RNA Northern blotting with a ChlI (SUL) probe.