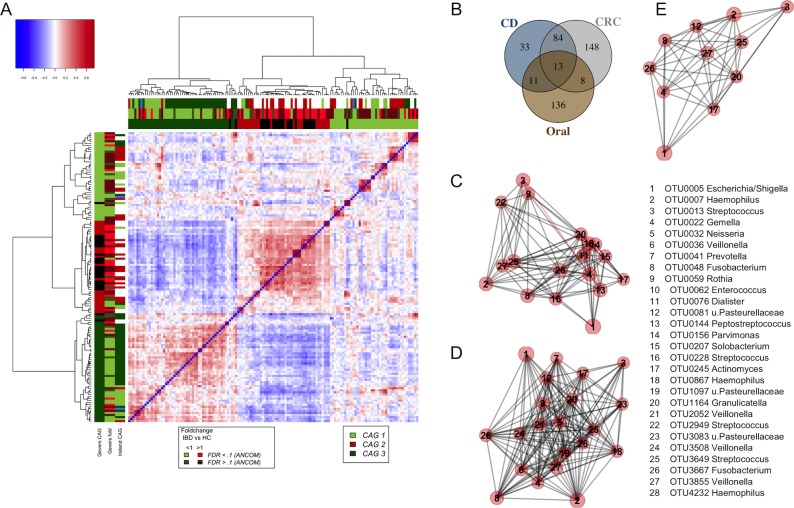
Figure 6.
Similarity of non-neoplastic and neoplastic colonic disease associated bacterial profiles. (A) Shown is the heatplot of the correlation values between OTUs associated with rectal tissue of children with and without CD.21 CAGs were defined on the basis of the clusters in the vertical or horizontal trees and named after their most notable characteristic. Column and row bars indicate bacterial CAGs (as per legend to the bottom right), fold change between individuals with CD and healthy controls (as per legend to the bottom left). Additionally, two row and column bars indicate the CAG in the Irish CRC cohort (figure 4A) and the fold change between individuals with CRCs/polyps and healthy controls. Legend top left: colour scale correlation coefficient. (B) Venn diagram of bacteria found in colorectal tissue of children with CD, colorectal tumours and oral swabs. (C–E) Network plots of bacterial OTUs found in both the oral cavity and different colonic tissue samples: (C) tumours (ON; 65 individuals with CRC and 2 polyps), (D) mucosa from children with CD (n=201) and (E) mucosa from healthy children (n=122). For each group of samples, the OTUs shared with the oral cavity was determined separately. The size of each node (OTU) correlates to the mean abundance of each OTU across all samples in each respective sample group. The width of each edge corresponds to the p value of the correlation between each respective node (lower p value, higher line width). The location of each node was determined by a PCoA of the correlation distance as described in Materials and Methods. Only nodes with at least one significant edge are shown. Legend to the right: genus-level classification using RDP reference, version 14 of OTU representative sequences. CAGs, coabundance groups; CD, Crohn’s disease; CRCs, colorectal cancer; OTUs, operational taxonomic units; PCoA, Principal Coordinates Analysis; RDP, Ribosomal Database Project.