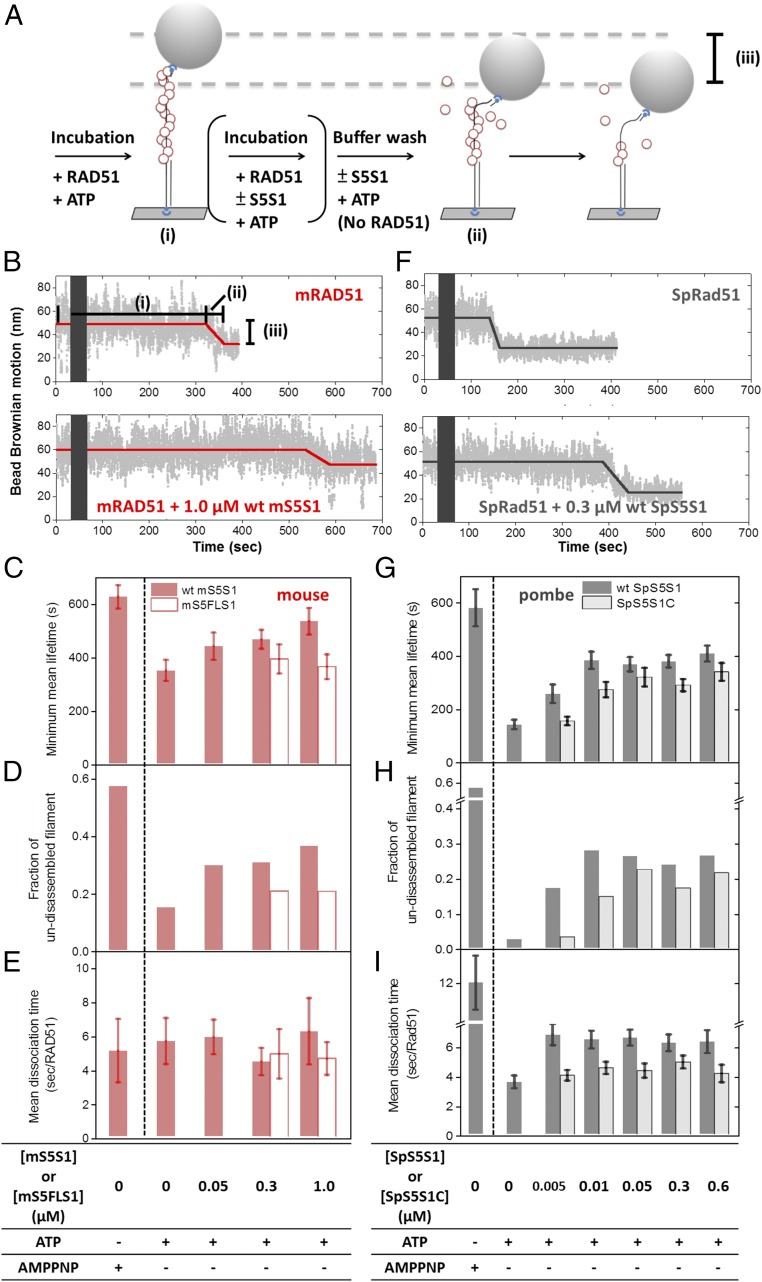
Fig. 5.
Nucleoprotein filament disassembly experiments showed that S5S1 prevents Rad51 filament disassembly. (A) Schematic illustration of nucleoprotein filament disassembly experiments using the TPM setup. (B) Representative bead BM time courses of mouse mRAD51 disassembly on (dT)135 DNA substrates without mS5S1 (Upper) and in the presence of 1.0 μM mS5S1 (Lower). mRAD51 filaments were preassembled in the presence of ATP. Dark gray bars stand for void time for extensive buffer wash to remove free mRAD51. A lifetime of the preassembled filament before the BM decrease dictates the mRAD51 disassembly kinetics. (C–E) Mean lifetime of mouse mRAD51 nucleoprotein filament (C), fraction of the undisassembled filament within 15 min (D), and mean dissociation time per mRAD51 (E) in the presence of various mS5S1 concentrations and nucleotide conditions. (F) Representative bead BM time courses of SpRad51 disassembly without SpS5S1 (Upper) and in the presence of 0.3 μM SpS5S1 (Lower). (G–I) Kinetic parameters for fission yeast. The fraction of undisassembled tethers is correlated with the mean lifetime of the SpRad51 filament in both species. Here, the N-terminal truncation S5S1C mutants (open bars) are deficient in DNA binding. All experiments were carried out at 2 mM ATP. Error bars are 1 SEM.