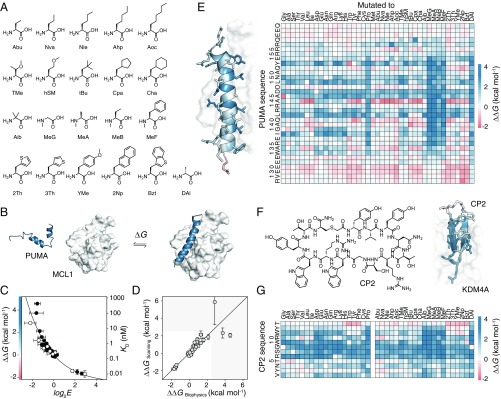
Fig. 2.
Nonproteinogenic deep mutational scanning of linear PUMA and cyclic CP2. (A) Test set of 21 nonproteinogenic amino acids used in this study. (B) PUMA (blue), an intrinsically disordered protein, folds to an α-helix upon binding with MCL1 (white) [Protein Data Bank (PDB) ID code 2ROC]. (C) Deep mutational scanning scores (log2E) report on free energy changes of binding upon mutation, ∆∆G. Proteinogenic (black) and nonproteinogenic mutants (white) overlay. Errors in log2E represent SDs of repeats of library binding and DNA recovery. (D) Leave-one-out cross-validation of empirical fit of ∆∆G and log2E showing agreement between calculated ∆∆G of the left-out mutant (∆∆GScanning) and expected experimental value (∆∆GBiophysics). Greater than 90% of PUMA mutant ∆∆G values fall within the white region. (E) ∆∆G for all mutants of PUMA; stabilizing (red) and destabilizing (blue). (Left) Average ∆∆G projected onto PUMA/MCL1 structure. (F) De novo macrocyclic peptide CP2 (Left). Average ∆∆G projected onto CP2/KDM4A structure (Right) (PDB ID code 5LY1). (G) ∆∆G for all mutants of CP2 binding to KDM4A.