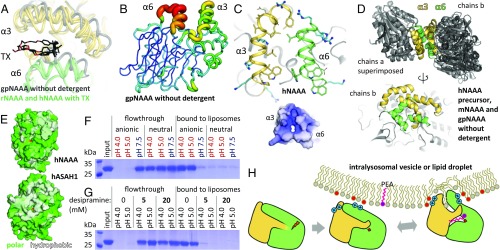
Fig. 5.
NAAA membrane interaction. (A) Conformational differences in helices α3 and α6 between gpNAAA bound to ARN726 in absence of detergent (gray), and other NAAA structures (each in yellow and green) in complex with inhibitors or product in presence of TX. (B) In gpNAAA, conformational flexibility of these helices is depicted by a plot of crystallographic B factors; a redder and thicker trace indicates higher disorder. (C) Helices α3 and α6 are mainly composed of hydrophobic residues (yellow and green sticks) and cationic side chains (blue sticks), resulting in an overall positive electrostatic surface potential at pH 4.5 (−5 kT/e, red to +5 kT/e, blue). (D) Superposition of crystallographic dimers (via chains a) from all crystal forms of NAAA obtained without detergent (Top), and alternate view of chains b, illustrating their variable orientations (Bottom). (E) In acid ceramidase (ASAH1, PDB ID code 5U7Z), a hydrophobic (white) bowl-shaped surface spans the α- and β-subunits. Residues were colored according to the Eisenberg hydrophobicity scale. (F) hNAAA was incubated with anionic or uncharged liposomes at various pH values. The vesicles were washed and pelleted, and bound protein was visualized by SDS/PAGE. (G) As in F, with the cationic amphiphilic drug desipramine added during incubation. (H) Proposed model of NAAA activation. After self-proteolysis in the lysosome, the enzyme electrostatically associates with intraluminal membranes. Contacts between lipids and its hydrophobic helices lead to a conformational change that exposes the substrate-binding site.