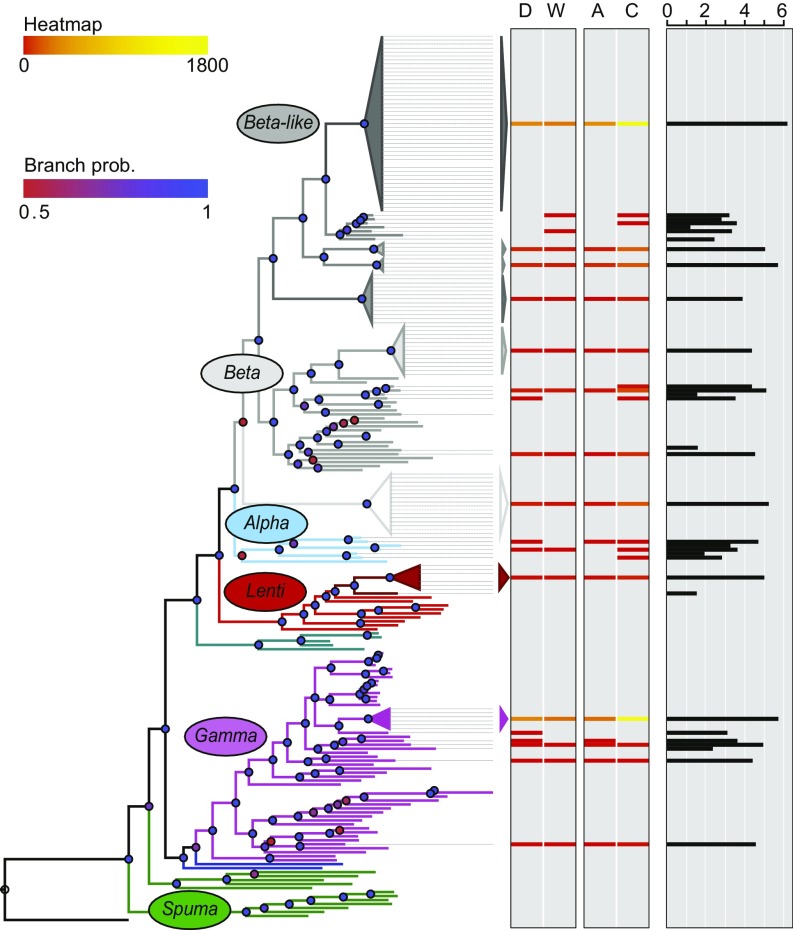
Fig. 4.
ERV phylogenetic association and spread among O. cuniculus sp. Retroviral gag- and pol-based tree derived from reference retroviruses and ERVs where colors indicate retrovirus-like nomenclature as previously described (4, 5). Dashed lines indicate ERVs identified in the rabbit reference genome assembly, oryCun2.0 (Dataset S1). The heatmap shows number of ERV insertions junctions in pairwise comparisons between domestic rabbit populations (D) and wild French rabbit populations (W), as well as between wild O. c. algirus populations (A) and wild O. c. cuniculus populations (C). The overall presence of candidate ERV insertion junctions across all O. cuniculus sp. populations is represented by the black histogram using log10 scale.