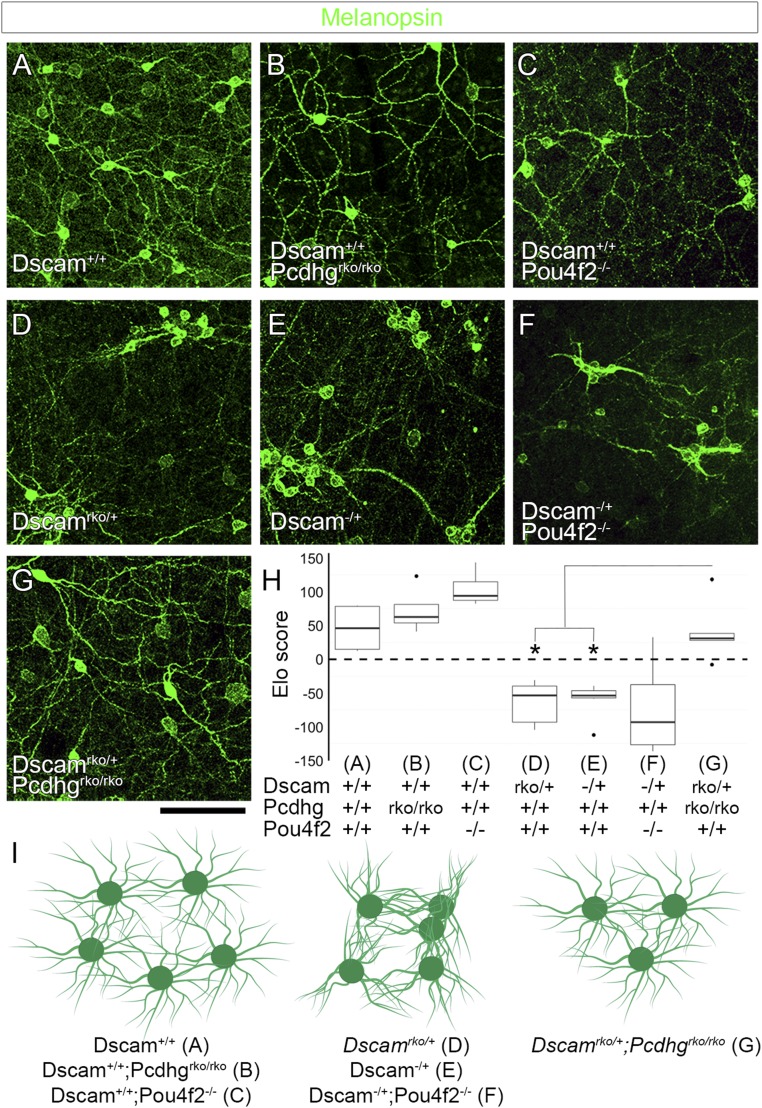
Fig. 6.
γ-Pcdh–mediated adhesion contributes to ipRGC fasciculation. (A) Confocal images from whole-mount retinas stained for melanopsin to label ipRGCs show normal spacing and dendritic coverage in wild-type animals. (B and C) Retina-specific deletion of Pcdhg or deletion of Pou4f2 reduces ipRGC cell number. (D and E) Retinas heterozygous for Dscam [Dscamrko/+ (D) or Dscam−/+ (E)] show significant ipRGC clustering and fasciculation. (F) This is not rescued by reducing cell number in Dscam−/+;Pou4f2−/− double mutants. (G) However, this loss of self-avoidance is rescued by reducing γ-Pcdh–mediated adhesion in Dscamrko/+;Pcdhgrko/rko double mutants. Image quantification is presented in H. * in H denotes P < 0.05 by pairwise Wilcoxon rank sum test compared to Dscamrko/+;Pcdhgrko/rko. These results are represented in I: Fasciculation is not observed in controls, Pcdhg mutants, or Pou4f2 mutants, although the latter two reduce cell number. Fasciculation is observed in Dscamrko/+, Dscam−/+, and Dscam−/+;Pou4f2−/−, despite changes in cell number, but these are rescued in Dscamrko/+;Pcdhgrko/rko double mutants. n = 6 retinas per genotype over two microscope fields of view per retina. Images of whole-retina quadrants were used for quantification (SI Appendix, Fig. S6). Box plots represent the median, first and third quartiles, range, and outliers. (Scale bar: 100 μm.)